Multiple recombinants in two dengue virus, serotype-2 isolates from patients from Oaxaca, Mexico
- PMID: 20003526
- PMCID: PMC2804599
- DOI: 10.1186/1471-2180-9-260
Multiple recombinants in two dengue virus, serotype-2 isolates from patients from Oaxaca, Mexico
Abstract
Background: Dengue (DEN) is a serious cause of mortality and morbidity in the world including Mexico, where the infection is endemic. One of the states with the highest rate of dengue cases is Oaxaca. The cause of DEN is a positive-sense RNA virus, the dengue virus (DENV) that evolves rapidly increasing its variability due to the absence of a repair mechanism that leads to approximately one mutational event per genome replication; which results in enhancement of viral adaptation, including the escape from host immune responses. Additionally, recombination may play a role in driving the evolution of DENV, which may potentially affect virulence and cause host tropism changes. Recombination in DENV has not been described in Mexican strains, neither has been described the relevance in virus evolution in an endemic state such as Oaxaca where the four serotypes of DENV are circulating.
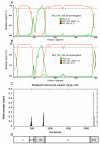
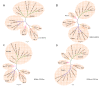
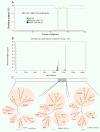
Results: To study whether there are isolates from Oaxaca having recombination, we obtained the sequence of 6 different isolates of DENV-2 Asian/American genotype from the outbreak 2005-6, one clone of the C(91)-prM-E-NS1(2400) structural genes, and 10 clones of the E gene from the isolate MEX_OAX_1656_05. Evidence of recombination was found by using different methods along with two softwares: RDP3 and GARD. The Oaxaca MEX_OAX_1656_05 and MEX_OAX_1038_05 isolates sequenced in this study were recombinant viruses that incorporate the genome sequence from the Cosmopolitan genotype. Furthermore, the clone of the E gene namely MEX_OAX_165607_05 from this study was also recombinant, incorporating genome sequence from the American genotype.
Conclusions: This is the first report of recombination in DENV-2 in Mexico. Given such a recombinant activity new genomic combinations were produced, this could play a significant role in the DENV evolution and must be considered as a potentially important mechanism generating genetic variation in this virus with serious implications for the vaccines and drugs formulation as occurs for other viruses like poliovirus, influenza and HIV.
Figures


Similar articles
-
Specific genetic markers for detecting subtypes of dengue virus serotype-2 in isolates from the states of Oaxaca and Veracruz, Mexico.BMC Microbiol. 2008 Jul 15;8:117. doi: 10.1186/1471-2180-8-117. BMC Microbiol. 2008. PMID: 18625078 Free PMC article.
-
Phylogenetic and recombinant analyses of complete coding sequences of DENV-1 from field-caught mosquitoes in Thailand.Virus Res. 2020 Sep;286:198041. doi: 10.1016/j.virusres.2020.198041. Epub 2020 Jun 1. Virus Res. 2020. PMID: 32497574
-
Molecular epidemiology of dengue viruses in southern China from 1978 to 2006.Virol J. 2011 Jun 26;8:322. doi: 10.1186/1743-422X-8-322. Virol J. 2011. PMID: 21703015 Free PMC article.
-
Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease.Infect Genet Evol. 2009 Jul;9(4):523-40. doi: 10.1016/j.meegid.2009.02.003. Epub 2009 Feb 13. Infect Genet Evol. 2009. PMID: 19460319 Free PMC article. Review.
-
Dengue--quo tu et quo vadis?Viruses. 2011 Sep;3(9):1562-608. doi: 10.3390/v3091562. Epub 2011 Sep 1. Viruses. 2011. PMID: 21994796 Free PMC article. Review.
Cited by
-
Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes.PeerJ. 2016 Aug 24;4:e2326. doi: 10.7717/peerj.2326. eCollection 2016. PeerJ. 2016. PMID: 27635316 Free PMC article.
-
Phylogenetic and evolutionary analysis of dengue virus serotypes circulating at the Colombian-Venezuelan border during 2015-2016 and 2018-2019.PLoS One. 2021 May 28;16(5):e0252379. doi: 10.1371/journal.pone.0252379. eCollection 2021. PLoS One. 2021. PMID: 34048474 Free PMC article.
-
Using bioinformatics tools for the discovery of Dengue RNA-dependent RNA polymerase inhibitors.PeerJ. 2018 Sep 25;6:e5068. doi: 10.7717/peerj.5068. eCollection 2018. PeerJ. 2018. PMID: 30280009 Free PMC article.
-
Prioritizing Water Security in the Management of Vector-Borne Diseases: Lessons From Oaxaca, Mexico.Geohealth. 2020 Mar 1;4(3):e2019GH000201. doi: 10.1029/2019GH000201. eCollection 2020 Mar. Geohealth. 2020. PMID: 32185244 Free PMC article.
-
The NS1 glycoprotein can generate dramatic antibody-enhanced dengue viral replication in normal out-bred mice resulting in lethal multi-organ disease.PLoS One. 2011;6(6):e21024. doi: 10.1371/journal.pone.0021024. Epub 2011 Jun 22. PLoS One. 2011. PMID: 21731643 Free PMC article.
References
-
- Gubler DJ, Meltzer M. Impact of dengue/dengue haemorrhagic fever on the developing world. Adv Virus Res. 1999;53:35–70. full_text. - PubMed
-
- Wang WK, Chao DY, Lin SR, King CC, Chang SC. Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. J Microbiol Immunol Infect. 2003;36:89–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

