Comprehensive molecular structure of the eukaryotic ribosome
- PMID: 20004163
- PMCID: PMC2814252
- DOI: 10.1016/j.str.2009.09.015
Comprehensive molecular structure of the eukaryotic ribosome
Abstract
Despite the emergence of a large number of X-ray crystallographic models of the bacterial 70S ribosome over the past decade, an accurate atomic model of the eukaryotic 80S ribosome is still not available. Eukaryotic ribosomes possess more ribosomal proteins and ribosomal RNA than do bacterial ribosomes, which are implicated in extraribosomal functions in the eukaryotic cells. By combining cryo-EM with RNA and protein homology modeling, we obtained an atomic model of the yeast 80S ribosome complete with all ribosomal RNA expansion segments and all ribosomal proteins for which a structural homolog can be identified. Mutation or deletion of 80S ribosomal proteins can abrogate maturation of the ribosome, leading to several human diseases. We have localized one such protein unique to eukaryotes, rpS19e, whose mutations are associated with Diamond-Blackfan anemia in humans. Additionally, we characterize crucial interactions between the dynamic stalk base of the ribosome with eukaryotic elongation factor 2.
Figures






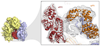
References
-
- Alkemar G, Nygard O. Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry. 2006;45:8067–8078. - PubMed
-
- Buchhaupt M, Meyer B, Kotter P, Entian KD. Genetic evidence for 18S rRNA binding and an Rps19p assembly function of yeast nucleolar protein Nep1p. Mol Genet Genomics. 2006;276:273–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083960/GM/NIGMS NIH HHS/United States
- P41 RR001219/RR/NCRR NIH HHS/United States
- U54 GM074945/GM/NIGMS NIH HHS/United States
- R01 GM053827/GM/NIGMS NIH HHS/United States
- GM55440/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM054762/GM/NIGMS NIH HHS/United States
- PN2 EY016525/EY/NEI NIH HHS/United States
- R37 GM029169/GM/NIGMS NIH HHS/United States
- R37 GM29169/GM/NIGMS NIH HHS/United States
- R01 GM055440/GM/NIGMS NIH HHS/United States
- R01 BM54762/BM/FDA HHS/United States
- P41 RR012255/RR/NCRR NIH HHS/United States
- GM53827/GM/NIGMS NIH HHS/United States
- RR12255/RR/NCRR NIH HHS/United States
- R01 BM083960/BM/FDA HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

