Identification of structural mechanisms of HIV-1 protease specificity using computational peptide docking: implications for drug resistance
- PMID: 20004167
- PMCID: PMC2806847
- DOI: 10.1016/j.str.2009.10.008
Identification of structural mechanisms of HIV-1 protease specificity using computational peptide docking: implications for drug resistance
Abstract
Drug-resistant mutations (DRMs) in HIV-1 protease are a major challenge to antiretroviral therapy. Protease-substrate interactions that are determined to be critical for native selectivity could serve as robust targets for drug design that are immune to DRMs. In order to identify the structural mechanisms of selectivity, we developed a peptide-docking algorithm to predict the atomic structure of protease-substrate complexes and applied it to a large and diverse set of cleavable and noncleavable peptides. Cleavable peptides showed significantly lower energies of interaction than noncleavable peptides with six protease active-site residues playing the most significant role in discrimination. Surprisingly, all six residues correspond to sequence positions associated with drug resistance mutations, demonstrating that the very residues that are responsible for native substrate specificity in HIV-1 protease are altered during its evolution to drug resistance, suggesting that drug resistance and substrate selectivity may share common mechanisms.
Figures






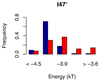






Similar articles
-
HIV-1 protease substrate-groove: Role in substrate recognition and inhibitor resistance.Biochimie. 2015 Nov;118:90-103. doi: 10.1016/j.biochi.2015.08.009. Epub 2015 Aug 20. Biochimie. 2015. PMID: 26300060
-
HIV-1 protease-substrate coevolution in nelfinavir resistance.J Virol. 2014 Jul;88(13):7145-54. doi: 10.1128/JVI.00266-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719428 Free PMC article.
-
Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.Protein Sci. 2012 Jul;21(7):1029-41. doi: 10.1002/pro.2086. Epub 2012 Jun 5. Protein Sci. 2012. PMID: 22549928 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
Cited by
-
Computational Selectivity Assessment of Protease Inhibitors against SARS-CoV-2.Int J Mol Sci. 2021 Feb 19;22(4):2065. doi: 10.3390/ijms22042065. Int J Mol Sci. 2021. PMID: 33669738 Free PMC article.
-
Unveiling the molecular activity of HIV towards the CD4: A study based on subtype C via docking and dynamics approach.J Genet Eng Biotechnol. 2025 Mar;23(1):100457. doi: 10.1016/j.jgeb.2025.100457. Epub 2025 Jan 16. J Genet Eng Biotechnol. 2025. PMID: 40074431 Free PMC article.
-
Interdependence of Inhibitor Recognition in HIV-1 Protease.J Chem Theory Comput. 2017 May 9;13(5):2300-2309. doi: 10.1021/acs.jctc.6b01262. Epub 2017 Apr 11. J Chem Theory Comput. 2017. PMID: 28358514 Free PMC article.
-
Drug Resistance Mechanism of M46I-Mutation-Induced Saquinavir Resistance in HIV-1 Protease Using Molecular Dynamics Simulation and Binding Energy Calculation.Viruses. 2022 Mar 28;14(4):697. doi: 10.3390/v14040697. Viruses. 2022. PMID: 35458427 Free PMC article.
-
Benchmarking and analysis of protein docking performance in Rosetta v3.2.PLoS One. 2011;6(8):e22477. doi: 10.1371/journal.pone.0022477. Epub 2011 Aug 2. PLoS One. 2011. PMID: 21829626 Free PMC article.
References
-
- Beck ZQ, Morris GM, Elder JH. Defining HIV-1 protease substrate selectivity. Current drug targets. 2002;2:37–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

