Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein
- PMID: 20004458
- PMCID: PMC2822108
- DOI: 10.1016/j.virol.2009.11.010
Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein
Abstract
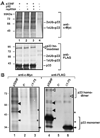
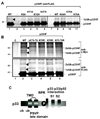
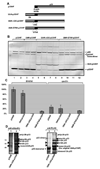
Post-translational modifications of viral replication proteins could be widespread phenomena during the replication of plus-stranded RNA viruses. In this article, we identify two lysines in the tombusvirus p33 replication co-factor involved in ubiquitination and show that the same lysines are also important for the p33 to interact with the host Vps23p ESCRT-I factor. We find that the interaction of p33 with Vps23p is also affected by a "late-domain"-like sequence in p33. The combined mutations of the two lysines and the late-domain-like sequences in p33 reduced replication of a replicon RNA of Tomato bushy stunt virus in yeast model host, in plant protoplasts, and plant leaves, suggesting that p33-Vps23p ESCRT protein interaction affects tombusvirus replication. Using ubiquitin-mimicking p33 chimeras, we demonstrate that high level of p33 ubiquitination is inhibitory for TBSV replication. These findings argue that optimal level of p33 ubiquitination plays a regulatory role during tombusvirus infections.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures




References
-
- Bamezai S, Tate S, Breslow E. Inhibition of ubiquitin-dependent proteolysis by des-Gly-Gly-ubiquitin: implications for the mechanism of polyubiquitin synthesis. Biochem Biophys Res Commun. 1989;162(1):89–94. - PubMed
-
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744(3):438–454. - PubMed
-
- Brigati C, Giacca M, Noonan DM, Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol Lett. 2003;220(1):57–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

