In silico and functional studies of the regulation of the glucocerebrosidase gene
- PMID: 20004604
- PMCID: PMC2827879
- DOI: 10.1016/j.ymgme.2009.10.189
In silico and functional studies of the regulation of the glucocerebrosidase gene
Abstract
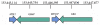
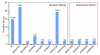
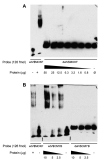
In Gaucher disease (GD), the inherited deficiency of glucocerebrosidase results in the accumulation of glucocerebroside within lysosomes. Although almost 300 mutations in the glucocerebrosidase gene (GBA) have been identified, the ability to predict phenotype from genotype is quite limited. In this study, we sought to examine potential GBA transcriptional regulatory elements for variants that contribute to phenotypic diversity. Specifically, we generated the genomic sequence for the orthologous genomic region ( approximately 39.4kb) encompassing GBA in eight non-human mammals. Computational comparisons of the resulting sequences, using human sequence as the reference, allowed the identification of multi-species conserved sequences (MCSs). Further analyses predicted the presence of two putative clusters of transcriptional regulatory elements upstream and downstream of GBA, containing five and three transcription factor-binding sites (TFBSs), respectively. A firefly luciferase (Fluc) reporter construct containing sequence flanking the GBA gene was used to test the functional consequences of altering these conserved sequences. The predicted TFBSs were individually altered by targeted mutagenesis, resulting in enhanced Fluc expression for one site and decreased expression for seven others sites. Gel-shift assays confirmed the loss of nuclear-protein binding for several of the mutated constructs. These identified conserved non-coding sequences flanking GBA could play a role in the transcriptional regulation of the gene contributing to the complexity underlying the phenotypic diversity seen in GD.
Published by Elsevier Inc.
Figures



Similar articles
-
Identification of three additional genes contiguous to the glucocerebrosidase locus on chromosome 1q21: implications for Gaucher disease.Genome Res. 1997 Oct;7(10):1020-6. doi: 10.1101/gr.7.10.1020. Genome Res. 1997. PMID: 9331372 Free PMC article.
-
Simple PCR amplification of the entire glucocerebrosidase gene (GBA) coding region for diagnostic sequence analysis.DNA Seq. 1998;8(6):349-56. doi: 10.3109/10425179809020896. DNA Seq. 1998. PMID: 10728820
-
Miglustat (NB-DNJ) works as a chaperone for mutated acid beta-glucosidase in cells transfected with several Gaucher disease mutations.Blood Cells Mol Dis. 2005 Sep-Oct;35(2):268-76. doi: 10.1016/j.bcmd.2005.05.007. Blood Cells Mol Dis. 2005. PMID: 16039881
-
Gaucher disease: complexity in a "simple" disorder.Mol Genet Metab. 2004 Sep-Oct;83(1-2):6-15. doi: 10.1016/j.ymgme.2004.08.015. Mol Genet Metab. 2004. PMID: 15464415 Review.
-
Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA).Hum Mutat. 2008 May;29(5):567-83. doi: 10.1002/humu.20676. Hum Mutat. 2008. PMID: 18338393 Review.
Cited by
-
Direct and indirect regulation of β-glucocerebrosidase by the transcription factors USF2 and ONECUT2.NPJ Parkinsons Dis. 2024 Oct 22;10(1):192. doi: 10.1038/s41531-024-00819-7. NPJ Parkinsons Dis. 2024. PMID: 39438499 Free PMC article.
-
Defective autophagy in Parkinson's disease: lessons from genetics.Mol Neurobiol. 2015 Feb;51(1):89-104. doi: 10.1007/s12035-014-8787-5. Epub 2014 Jul 4. Mol Neurobiol. 2015. PMID: 24990317 Review.
-
Exploring genetic modifiers of Gaucher disease: The next horizon.Hum Mutat. 2018 Dec;39(12):1739-1751. doi: 10.1002/humu.23611. Epub 2018 Sep 11. Hum Mutat. 2018. PMID: 30098107 Free PMC article. Review.
-
Identification of recombinant alleles using quantitative real-time PCR implications for Gaucher disease.J Mol Diagn. 2011 Jul;13(4):401-5. doi: 10.1016/j.jmoldx.2011.02.005. J Mol Diagn. 2011. PMID: 21704274 Free PMC article.
-
Glucocerebrosidase is shaking up the synucleinopathies.Brain. 2014 May;137(Pt 5):1304-22. doi: 10.1093/brain/awu002. Epub 2014 Feb 14. Brain. 2014. PMID: 24531622 Free PMC article. Review.
References
-
- Beutler E, Grabowski G. Gaucher Disease. In: Scriver CBA, Sly W, Valle D, editors. The Metabolic & Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 3635–3668.
-
- Sidransky E. Gaucher disease: complexity in a “simple” disorder. Molecular genetics and metabolism. 2004;83:6–15. - PubMed
-
- Goker-Alpan O, Schiffmann R, Park JK, et al. Phenotypic continuum in neuronopathic Gaucher disease: an intermediate phenotype between type 2 and type 3. The Journal of pediatrics. 2003;143:273–276. - PubMed
-
- Hruska KS, LaMarca ME, Scott CR, et al. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Human mutation. 2008;29:567–583. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

