The biological coherence of human phenome databases
- PMID: 20004759
- PMCID: PMC2790572
- DOI: 10.1016/j.ajhg.2009.10.026
The biological coherence of human phenome databases
Abstract
Disease networks are increasingly explored as a complement to networks centered around interactions between genes and proteins. The quality of disease networks is heavily dependent on the amount and quality of phenotype information in phenotype databases of human genetic diseases. We explored which aspects of phenotype database architecture and content best reflect the underlying biology of disease. We used the OMIM-based HPO, Orphanet, and POSSUM phenotype databases for this purpose and devised a biological coherence score based on the sharing of gene ontology annotation to investigate the degree to which phenotype similarity in these databases reflects related pathobiology. Our analyses support the notion that a fine-grained phenotype ontology enhances the accuracy of phenome representation. In addition, we find that the OMIM database that is most used by the human genetics community is heavily underannotated. We show that this problem can easily be overcome by simply adding data available in the POSSUM database to improve OMIM phenotype representations in the HPO. Also, we find that the use of feature frequency estimates--currently implemented only in the Orphanet database--significantly improves the quality of the phenome representation. Our data suggest that there is much to be gained by improving human phenome databases and that some of the measures needed to achieve this are relatively easy to implement. More generally, we propose that curation and more systematic annotation of human phenome databases can greatly improve the power of the phenotype for genetic disease analysis.
Figures
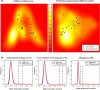
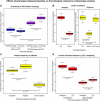
References
-
- Oti M., Brunner H.G. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. - PubMed
-
- Oti M., Huynen M.A., Brunner H.G. Phenome connections. Trends Genet. 2008;24:103–106. - PubMed
-
- Brunner H.G., van Driel M.A. From syndrome families to functional genomics. Nat. Rev. Genet. 2004;5:545–551. - PubMed
-
- Freudenberg J., Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18(Suppl 2):S110–S115. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

