Dosage-dependent severity of the phenotype in patients with mental retardation due to a recurrent copy-number gain at Xq28 mediated by an unusual recombination
- PMID: 20004760
- PMCID: PMC2790561
- DOI: 10.1016/j.ajhg.2009.10.019
Dosage-dependent severity of the phenotype in patients with mental retardation due to a recurrent copy-number gain at Xq28 mediated by an unusual recombination
Abstract
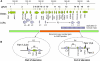
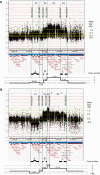
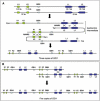
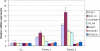
We report on the identification of a 0.3 Mb inherited recurrent but variable copy-number gain at Xq28 in affected males of four unrelated families with X-linked mental retardation (MR). All aberrations segregate with the disease in the families, and the carrier mothers show nonrandom X chromosome inactivation. Tiling Xq28-region-specific oligo array revealed that all aberrations start at the beginning of the low copy repeat LCR-K1, at position 153.20 Mb, and end just distal to LCR-L2, at 153.54 Mb. The copy-number gain always includes 18 annotated genes, of which RPL10, ATP6AP1 and GDI1 are highly expressed in brain. From these, GDI1 is the most likely candidate gene. Its copy number correlates with the severity of clinical features, because it is duplicated in one family with nonsyndromic moderate MR, is triplicated in males from two families with mild MR and additional features, and is present in five copies in a fourth family with a severe syndromic form of MR. Moreover, expression analysis revealed copy-number-dependent increased mRNA levels in affected patients compared to control individuals. Interestingly, analysis of the breakpoint regions suggests a recombination mechanism that involves two adjacent but different sets of low copy repeats. Taken together, our data strongly suggest that an increased expression of GDI1 results in impaired cognition in a dosage-dependent manner. Moreover, these data also imply that a copy-number gain of an individual gene present in the larger genomic aberration that leads to the severe MECP2 duplication syndrome can of itself result in a clinical phenotype as well.
Figures


Comment in
-
The LCR at the IKBKG locus is prone to recombine.Am J Hum Genet. 2010 Apr 9;86(4):650-2; author reply 652-3. doi: 10.1016/j.ajhg.2009.12.019. Am J Hum Genet. 2010. PMID: 20380930 Free PMC article. No abstract available.
References
-
- Froyen G., Van Esch H., Bauters M., Hollanders K., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: Important role for increased gene dosage of XLMR genes. Hum. Mutat. 2007;28:1034–1042. - PubMed
-
- Froyen G., Corbett M., Vandewalle J., Jarvela I., Lawrence O., Meldrum C., Bauters M., Govaerts K., Vandeleur L., Van Esch H. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am. J. Hum. Genet. 2008;82:432–443. - PMC - PubMed
-
- Van Esch H., Bauters M., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005;77:442–453. - PMC - PubMed
-
- Bauters M., Van Esch H., Friez M.J., Boespflug-Tanguy O., Zenker M., Vianna-Morgante A.M., Rosenberg C., Ignatius J., Raynaud M., Hollanders K. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–858. - PMC - PubMed
-
- Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

