Optimizing protein stability in vivo
- PMID: 20005848
- PMCID: PMC2818778
- DOI: 10.1016/j.molcel.2009.11.022
Optimizing protein stability in vivo
Abstract
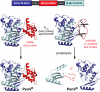
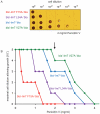
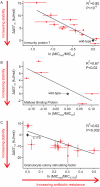
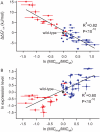
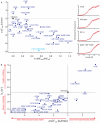
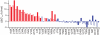
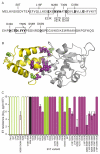
Identifying mutations that stabilize proteins is challenging because most substitutions are destabilizing. In addition to being of immense practical utility, the ability to evolve protein stability in vivo may indicate how evolution has formed today's protein sequences. Here we describe a genetic selection that directly links the in vivo stability of proteins to antibiotic resistance. It allows the identification of stabilizing mutations within proteins. The large majority of mutants selected for improved antibiotic resistance are stabilized both thermodynamically and kinetically, indicating that similar principles govern stability in vivo and in vitro. The approach requires no prior structural or functional knowledge and allows selection for stability without a need to maintain function. Mutations that enhance thermodynamic stability of the protein Im7 map overwhelmingly to surface residues involved in binding to colicin E7, showing how the evolutionary pressures that drive Im7-E7 complex formation have compromised the stability of the isolated Im7 protein.
Figures
Comment in
-
Evolving protein stability through genetic selection.Mol Cell. 2009 Dec 11;36(5):730-1. doi: 10.1016/j.molcel.2009.11.023. Mol Cell. 2009. PMID: 20005835
References
-
- Bishop B, Koay DC, Sartorelli AC, Regan L. Reengineering granulocyte colony-stimulating factor for enhanced stability. J. Biol. Chem. 2001;276:33465–33470. - PubMed
-
- Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 2002;9:209–216. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

