Assembly of the Sos1-Grb2-Gab1 ternary signaling complex is under allosteric control
- PMID: 20005866
- PMCID: PMC2819574
- DOI: 10.1016/j.abb.2009.12.011
Assembly of the Sos1-Grb2-Gab1 ternary signaling complex is under allosteric control
Abstract
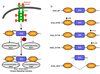
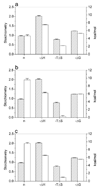
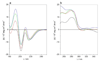
Allostery has evolved as a form of local communication between interacting protein partners allowing them to quickly sense changes in their immediate vicinity in response to external cues. Herein, using isothermal titration calorimetry (ITC) in conjunction with circular dichroism (CD) and macromolecular modeling (MM), we show that the binding of Grb2 adaptor--a key signaling molecule involved in the activation of Ras GTPase--to its downstream partners Sos1 guanine nucleotide exchange factor and Gab1 docker is under tight allosteric regulation. Specifically, our findings reveal that the binding of one molecule of Sos1 to the nSH3 domain allosterically induces a conformational change within Grb2 such that the loading of a second molecule of Sos1 onto the cSH3 domain is blocked and, in so doing, allows Gab1 access to the cSH3 domain in an exclusively non-competitive manner to generate the Sos1-Grb2-Gab1 ternary signaling complex.
2009 Elsevier Inc. All rights reserved.
Figures



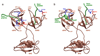
References
-
- Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. Nature. 1958;181:662–666. - PubMed
-
- Monod J, Changeux JP, Jacob F. J Mol Biol. 1963;6:306–329. - PubMed
-
- Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88–118. - PubMed
-
- Kuriyan J, Eisenberg D. Nature. 2007;450:983–990. - PubMed
-
- Chardin P, Cussac D, Maignan S, Ducruix A. FEBS Lett. 1995;369:47–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

