Nucleosome assembly depends on the torsion in the DNA molecule: a magnetic tweezers study
- PMID: 20006952
- PMCID: PMC2793352
- DOI: 10.1016/j.bpj.2009.09.032
Nucleosome assembly depends on the torsion in the DNA molecule: a magnetic tweezers study
Abstract
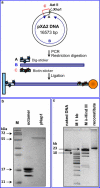
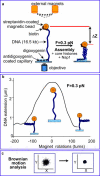
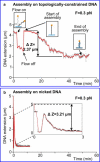
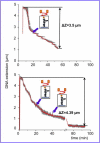
We have used magnetic tweezers to study nucleosome assembly on topologically constrained DNA molecules. Assembly was achieved using chicken erythrocyte core histones and histone chaperone protein Nap1 under constant low force. We have observed only partial assembly when the DNA was topologically constrained and much more complete assembly on unconstrained (nicked) DNA tethers. To verify our hypothesis that the lack of full nucleosome assembly on topologically constrained tethers was due to compensatory accumulation of positive supercoiling in the rest of the template, we carried out experiments in which we mechanically relieved the positive supercoiling by rotating the external magnetic field at certain time points of the assembly process. Indeed, such rotation did lead to the same nucleosome saturation level as in the case of nicked tethers. We conclude that levels of positive supercoiling in the range of 0.025-0.051 (most probably in the form of twist) stall the nucleosome assembly process.
Figures
References
-
- van Holde K.E. Springer-Verlag; New York: 1989. Chromatin.
-
- Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. - PubMed
-
- Zlatanova J., Leuba S.H. Elsevier; Amsterdam, New York: 2004. Chromatin Structure and Dynamics: State-of-the-Art.
-
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

