Force-driven separation of short double-stranded DNA
- PMID: 20006953
- PMCID: PMC2793367
- DOI: 10.1016/j.bpj.2009.09.040
Force-driven separation of short double-stranded DNA
Abstract
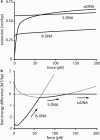
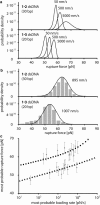
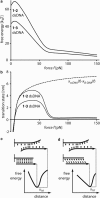
Short double-stranded DNA is used in a variety of nanotechnological applications, and for many of them, it is important to know for which forces and which force loading rates the DNA duplex remains stable. In this work, we develop a theoretical model that describes the force-dependent dissociation rate for DNA duplexes tens of basepairs long under tension along their axes ("shear geometry"). Explicitly, we set up a three-state equilibrium model and apply the canonical transition state theory to calculate the kinetic rates for strand unpairing and the rupture-force distribution as a function of the separation velocity of the end-to-end distance. Theory is in excellent agreement with actual single-molecule force spectroscopy results and even allows for the prediction of the rupture-force distribution for a given DNA duplex sequence and separation velocity. We further show that for describing double-stranded DNA separation kinetics, our model is a significant refinement of the conventionally used Bell-Evans model.
Figures


Similar articles
-
Force and kinetic barriers to unzipping of the DNA double helix.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8608-13. doi: 10.1073/pnas.151257598. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447279 Free PMC article.
-
Force measurements reveal how small binders perturb the dissociation mechanisms of DNA duplex sequences.Nanoscale. 2016 Jun 2;8(22):11718-26. doi: 10.1039/c6nr02201d. Nanoscale. 2016. PMID: 27221618
-
Force-Induced Rupture of a DNA Duplex: From Fundamentals to Force Sensors.ACS Nano. 2015 Dec 22;9(12):11993-2003. doi: 10.1021/acsnano.5b04726. Epub 2015 Nov 30. ACS Nano. 2015. PMID: 26575598
-
Force and kinetic barriers to initiation of DNA unzipping.Phys Rev E Stat Nonlin Soft Matter Phys. 2002 Apr;65(4 Pt 1):041907. doi: 10.1103/PhysRevE.65.041907. Epub 2002 Mar 28. Phys Rev E Stat Nonlin Soft Matter Phys. 2002. PMID: 12005873
-
Molecular force balance measurements reveal that double-stranded DNA unbinds under force in rate-dependent pathways.Biophys J. 2008 Jun;94(12):4766-74. doi: 10.1529/biophysj.107.125427. Epub 2008 Mar 13. Biophys J. 2008. PMID: 18339733 Free PMC article.
Cited by
-
Weak tension accelerates hybridization and dehybridization of short oligonucleotides.Nucleic Acids Res. 2023 Apr 24;51(7):3030-3040. doi: 10.1093/nar/gkad118. Nucleic Acids Res. 2023. PMID: 36869666 Free PMC article.
-
Improving single molecule force spectroscopy through automated real-time data collection and quantification of experimental conditions.Ultramicroscopy. 2014 Jan;136:7-14. doi: 10.1016/j.ultramic.2013.07.020. Epub 2013 Aug 7. Ultramicroscopy. 2014. PMID: 24001740 Free PMC article.
-
Increasing valence pushes DNA nanostar networks to the isostatic point.Proc Natl Acad Sci U S A. 2019 Apr 9;116(15):7238-7243. doi: 10.1073/pnas.1819683116. Epub 2019 Mar 26. Proc Natl Acad Sci U S A. 2019. PMID: 30914457 Free PMC article.
-
A three-state model with loop entropy for the overstretching transition of DNA.Biophys J. 2010 Jul 21;99(2):578-87. doi: 10.1016/j.bpj.2010.04.046. Biophys J. 2010. PMID: 20643077 Free PMC article.
-
Single-cell mechanogenetics using monovalent magnetoplasmonic nanoparticles.Nat Protoc. 2017 Sep;12(9):1871-1889. doi: 10.1038/nprot.2017.071. Epub 2017 Aug 17. Nat Protoc. 2017. PMID: 28817122 Free PMC article.
References
-
- Seeman N. DNA in a material world. Nature. 2003;421:427–431. - PubMed
-
- Martin M. DNA handles for single molecule experiments. Single Mol. 2000;1:139–144.
-
- Yan H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. - PubMed
-
- Cohen J.D., Sadowski J.P., Dervan P.B. Addressing single molecules on DNA nanostructures. Angew. Chem. Int. Ed. 2007;46:7956–7959. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

