5-methylcytosine in RNA: detection, enzymatic formation and biological functions
- PMID: 20007150
- PMCID: PMC2836557
- DOI: 10.1093/nar/gkp1117
5-methylcytosine in RNA: detection, enzymatic formation and biological functions
Abstract
The nucleobase modification 5-methylcytosine (m(5)C) is widespread both in DNA and different cellular RNAs. The functions and enzymatic mechanisms of DNA m(5)C-methylation were extensively studied during the last decades. However, the location, the mechanism of formation and the cellular function(s) of the same modified nucleobase in RNA still remain to be elucidated. The recent development of a bisulfite sequencing approach for efficient m(5)C localization in various RNA molecules puts ribo-m(5)C in a highly privileged position as one of the few RNA modifications whose detection is amenable to PCR-based amplification and sequencing methods. Additional progress in the field also includes the characterization of several specific RNA methyltransferase enzymes in various organisms, and the discovery of a new and unexpected link between DNA and RNA m(5)C-methylation. Numerous putative RNA:m(5)C-MTases have now been identified and are awaiting characterization, including the identification of their RNA substrates and their related cellular functions. In order to bring these recent exciting developments into perspective, this review provides an ordered overview of the detection methods for RNA methylation, of the biochemistry, enzymology and molecular biology of the corresponding modification enzymes, and discusses perspectives for the emerging biological functions of these enzymes.
Figures

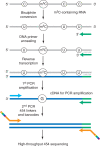
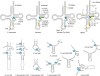


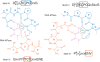
References
-
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948;175:315–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

