An intermolecular RNA triplex provides insight into structural determinants for the pseudoknot stimulator of -1 ribosomal frameshifting
- PMID: 20007152
- PMCID: PMC2836554
- DOI: 10.1093/nar/gkp1107
An intermolecular RNA triplex provides insight into structural determinants for the pseudoknot stimulator of -1 ribosomal frameshifting
Abstract
An efficient -1 programmed ribosomal frameshifting (PRF) signal requires an RNA slippery sequence and a downstream RNA stimulator, and the hairpin-type pseudoknot is the most common stimulator. However, a pseudoknot is not sufficient to promote -1 PRF. hTPK-DU177, a pseudoknot derived from human telomerase RNA, shares structural similarities with several -1 PRF pseudoknots and is used to dissect the roles of distinct structural features in the stimulator of -1 PRF. Structure-based mutagenesis on hTPK-DU177 reveals that the -1 PRF efficiency of this stimulator can be modulated by sequential removal of base-triple interactions surrounding the helical junction. Further analysis of the junction-flanking base triples indicates that specific stem-loop interactions and their relative positions to the helical junction play crucial roles for the -1 PRF activity of this pseudoknot. Intriguingly, a bimolecular pseudoknot approach based on hTPK-DU177 reveals that continuing triplex structure spanning the helical junction, lacking one of the loop-closure features embedded in pseudoknot topology, can stimulate -1 PRF. Therefore, the triplex structure is an essential determinant for the DU177 pseudoknot to stimulate -1 PRF. Furthermore, it suggests that -1 PRF, induced by an in-trans RNA via specific base-triple interactions with messenger RNAs, can be a plausible regulatory function for non-coding RNAs.
Figures

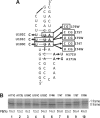


Similar articles
-
Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting.Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12706-11. doi: 10.1073/pnas.0905046106. Epub 2009 Jul 23. Proc Natl Acad Sci U S A. 2009. PMID: 19628688 Free PMC article.
-
A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated -1 ribosomal frameshifting.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12694-9. doi: 10.1073/pnas.0506166102. Epub 2005 Aug 25. Proc Natl Acad Sci U S A. 2005. PMID: 16123125 Free PMC article.
-
Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: a mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site.RNA. 2002 Aug;8(8):981-96. doi: 10.1017/s1355838202024044. RNA. 2002. PMID: 12212853 Free PMC article.
-
Regulators of Viral Frameshifting: More Than RNA Influences Translation Events.Annu Rev Virol. 2020 Sep 29;7(1):219-238. doi: 10.1146/annurev-virology-012120-101548. Epub 2020 Jun 29. Annu Rev Virol. 2020. PMID: 32600156 Free PMC article. Review.
-
Frameshifting RNA pseudoknots: structure and mechanism.Virus Res. 2009 Feb;139(2):193-208. doi: 10.1016/j.virusres.2008.06.008. Epub 2008 Jul 25. Virus Res. 2009. PMID: 18621088 Free PMC article. Review.
Cited by
-
Premature translation termination mediated non-ER stress induced ATF6 activation by a ligand-dependent ribosomal frameshifting circuit.Nucleic Acids Res. 2022 May 20;50(9):5369-5383. doi: 10.1093/nar/gkac257. Nucleic Acids Res. 2022. PMID: 35511080 Free PMC article.
-
Sensitive and label-free biosensing of RNA with predicted secondary structures by a triplex affinity capture method.Nucleic Acids Res. 2012 Apr;40(8):e56. doi: 10.1093/nar/gkr1304. Epub 2012 Jan 12. Nucleic Acids Res. 2012. PMID: 22241768 Free PMC article.
-
Ensemble simulations: folding, unfolding and misfolding of a high-efficiency frameshifting RNA pseudoknot.Nucleic Acids Res. 2017 May 5;45(8):4893-4904. doi: 10.1093/nar/gkx012. Nucleic Acids Res. 2017. PMID: 28115636 Free PMC article.
-
Coordination among tertiary base pairs results in an efficient frameshift-stimulating RNA pseudoknot.Nucleic Acids Res. 2017 Jun 2;45(10):6011-6022. doi: 10.1093/nar/gkx134. Nucleic Acids Res. 2017. PMID: 28334864 Free PMC article.
-
Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells.Nucleic Acids Res. 2014 Dec 16;42(22):14070-82. doi: 10.1093/nar/gku1233. Epub 2014 Nov 20. Nucleic Acids Res. 2014. PMID: 25414357 Free PMC article.
References
-
- Gesteland RF, Atkins JF. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. - PubMed
-
- Jacks T, Varmus HE. Expression of the Rous sarcoma virus polgene by ribosomal frameshifting. Science. 1985;230:1237–1242. - PubMed
-
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. - PubMed

