Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction
- PMID: 20007265
- PMCID: PMC2812397
- DOI: 10.1128/JVI.01902-09
Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction
Abstract
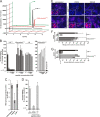
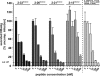
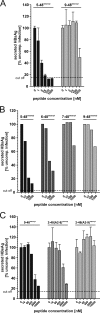
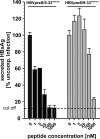
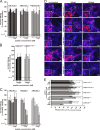
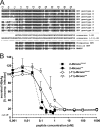
Previous studies showed that the N-terminal 75 amino acids of the pre-S1 domain of the hepatitis B virus (HBV) L protein are essential for HBV and hepatitis delta virus (HDV) infectivity. Consistently, synthetic lipopeptides encompassing this sequence or only parts of it efficiently block HBV and HDV infection, presumably through specific interference with a cellular receptor. Crucial for both virus infectivity and the inhibitory activity of the peptides are N-terminal myristoylation and a highly conserved motif within the N-terminal 48 amino acids. To refine the sequence requirements, we synthesized a series of HBV pre-S1 peptides containing deletions, point mutations, d-amino acid exchanges, or genotype-specific sequence permutations. Using the HepaRG cell line and a genotype D-derived virus, we determined the specific inhibitory activities of the peptides and found that (i) lipopeptides with an artificial consensus sequence inhibit HBV genotype D infection more potently than the corresponding genotype D peptides; (ii) point mutations, d-amino acid exchanges, or deletions introduced into the highly conserved part of the pre-S1 domain result in an almost complete loss of activity; and (iii) the flanking sequences comprising amino acids 2 to 8, 16 to 20, and, to a less pronounced extent, 34 to 48 gradually increase the inhibitory activity, while amino acids 21 to 33 behave indifferently. Taken together, our data suggest that HBV pre-S1-mediated receptor interference and, thus, HBV receptor recognition form a highly specific process. It requires an N-terminal acyl moiety and a highly conserved sequence that is present in primate but not rodent or avian hepadnaviruses, indicating different entry pathways for the different family members.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

