During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes
- PMID: 20007274
- PMCID: PMC2812404
- DOI: 10.1128/JVI.01934-09
During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes
Abstract
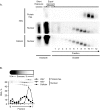
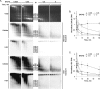
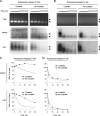
The genomes of herpes simplex virus type 1 (HSV-1) are regularly chromatinized during latency such that their digestion with micrococcal nuclease (MCN) releases nucleosome-sized DNA fragments. In lytically infected cells, in contrast, MCN releases HSV-1 DNA in primarily heterogeneously sized fragments. Consistently, only a small percentage of this HSV-1 DNA coimmunoprecipitates with histones. Most current models propose that histones associate with HSV-1 DNA during lytic infections at low occupancy. However, histone modification or occupation is also proposed to regulate HSV-1 transcription. It remains unclear how the histones associated with a small percentage of HSV-1 DNA may regulate transcription globally. Moreover, the physical properties of the complexes containing histones and HSV-1 DNA are unknown. We evaluated the HSV-1 DNA-containing complexes at 5 h after (lytic) infection by biochemical fractionations. Nuclear HSV-1 DNA did not fractionate as protein-free HSV-1 DNA but as DNA in cellular nucleosomes. Moreover, MCN released HSV-1 DNA in complexes that fractionate as cellular mono- and dinucleosomes by centrifugation followed by sucrose gradients and size-exclusion chromatography. The HSV-1 DNA in such complexes was protected to heterogeneous sizes and was more accessible to MCN than DNA in most cellular chromatin. Using a modified MCN digestion to trap unstable digestion intermediates, HSV-1 DNA was quantitatively recovered in discrete mono- to polynucleosome sizes in complexes fractionating as cellular mono- to polynucleosomes. The HSV-1 DNAs in complexes fractionating as mono- to dinucleosomes were stabilized by cross-linking. Therefore, most HSV-1 DNA forms particularly unstable nucleosome-like complexes at 5 h of lytic infection.
Figures

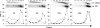
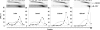


References
-
- Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063-2068. - PMC - PubMed
-
- Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451-463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

