Influenza C virus NS1 protein upregulates the splicing of viral mRNAs
- PMID: 20007279
- PMCID: PMC2812400
- DOI: 10.1128/JVI.01627-09
Influenza C virus NS1 protein upregulates the splicing of viral mRNAs
Abstract
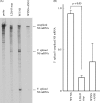
Pre-mRNAs of the influenza A virus M and NS genes are poorly spliced in virus-infected cells. By contrast, in influenza C virus-infected cells, the predominant transcript from the M gene is spliced mRNA. The present study was performed to investigate the mechanism by which influenza C virus M gene-specific mRNA (M mRNA) is readily spliced. The ratio of M1 encoded by a spliced M mRNA to CM2 encoded by an unspliced M mRNA in influenza C virus-infected cells was about 10 times larger than that in M gene-transfected cells, suggesting that a viral protein(s) other than M gene translational products facilitates viral mRNA splicing. RNase protection assays showed that the splicing of M mRNA in infected cells was much higher than that in M gene-transfected cells. The unspliced and spliced mRNAs of the influenza C virus NS gene encode two nonstructural (NS) proteins, NS1(C/NS1) and NS2(C/NS2), respectively. The introduction of premature translational termination into the NS gene, which blocked the synthesis of the C/NS1 and C/NS2 proteins, drastically reduced the splicing of NS mRNA, raising the possibility that C/NS1 or C/NS2 enhances viral mRNA splicing. The splicing of influenza C virus M mRNA was increased by coexpression of C/NS1, whereas it was reduced by coexpression of the influenza A virus NS1 protein (A/NS1). The splicing of influenza A virus M mRNA was also increased by coexpression of C/NS1, though it was inhibited by that of A/NS1. These results suggest that influenza C virus NS1, but not A/NS1, can upregulate viral mRNA splicing.
Figures
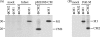








References
-
- Alamgir, A. S. M., Y. Matsuzaki, S. Hongo, E. Tsuchiya, K. Sugawara, Y. Muraki, and K. Nakamura. 2000. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J. Gen. Virol. 81:1933-1940. - PubMed
-
- Betakova, T., and A. J. Hay. 2007. Evidence that the CM2 protein of influenza C virus can modify the pH of the exocytic pathway of transfected cells. J. Gen. Virol. 88:2291-2296. - PubMed
-
- Bukrinskaya, A. G., N. K. Vorunova, G. V. Kornilayeva, R. A. Narmanbetova, and A. J. Hay. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49-59. - PubMed
-
- Ciampor, F., C. A. Thompson, S. Grambas, and A. J. Hay. 1992. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 22:247-258. - PubMed
-
- Enami, M., G. Sharma, C. Benham, and P. Palese. 1991. An influenza virus containing nine different RNA segments. Virology 185:291-298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

