A single tyrosine in the severe acute respiratory syndrome coronavirus membrane protein cytoplasmic tail is important for efficient interaction with spike protein
- PMID: 20007283
- PMCID: PMC2812384
- DOI: 10.1128/JVI.02458-09
A single tyrosine in the severe acute respiratory syndrome coronavirus membrane protein cytoplasmic tail is important for efficient interaction with spike protein
Abstract
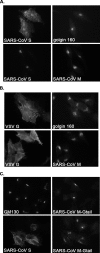
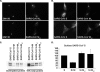
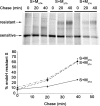
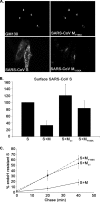
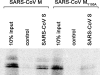
Severe acute respiratory syndrome coronavirus (SARS-CoV) encodes 3 major envelope proteins: spike (S), membrane (M), and envelope (E). Previous work identified a dibasic endoplasmic reticulum retrieval signal in the cytoplasmic tail of SARS-CoV S that promotes efficient interaction with SARS-CoV M. The dibasic signal was shown to be important for concentrating S near the virus assembly site rather than for direct interaction with M. Here, we investigated the sequence requirements of the SARS-CoV M protein that are necessary for interaction with SARS-CoV S. The SARS-CoV M tail was shown to be necessary for S localization in the Golgi region when the proteins were exogenously coexpressed in cells. This was specific, since SARS-CoV M did not retain an unrelated glycoprotein in the Golgi. Importantly, we found that an essential tyrosine residue in the SARS-CoV M cytoplasmic tail, Y(195), was important for S-M interaction. When Y(195) was mutated to alanine, M(Y195A) no longer retained S intracellularly at the Golgi. Unlike wild-type M, M(Y195A) did not reduce the amount of SARS-CoV S carbohydrate processing or surface levels when the two proteins were coexpressed. Mutating Y(195) also disrupted SARS-CoV S-M interaction in vitro. These results suggest that Y(195) is necessary for efficient SARS-CoV S-M interaction and, thus, has a significant involvement in assembly of infectious virus.
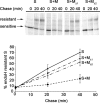
Figures


References
-
- Armstrong, J., S. Patel, and P. Riddle. 1990. Lysosomal sorting mutants of coronavirus E1 protein, a Golgi membrane protein. J. Cell Sci. 95(Pt. 2):191-197. - PubMed
-
- Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

