Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase
- PMID: 20007320
- PMCID: PMC2820805
- DOI: 10.1074/jbc.M109.065516
Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase
Abstract
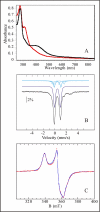
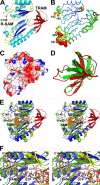
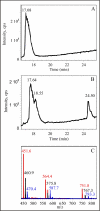
Post-translational modifications of ribosomal proteins are important for the accuracy of the decoding machinery. A recent in vivo study has shown that the rimO gene is involved in generation of the 3-methylthio derivative of residue Asp-89 in ribosomal protein S12 (Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1826-1831). This reaction is formally identical to that catalyzed by MiaB on the C2 of adenosine 37 near the anticodon of several tRNAs. We present spectroscopic evidence that Thermotoga maritima RimO, like MiaB, contains two [4Fe-4S] centers, one presumably bound to three invariant cysteines in the central radical S-adenosylmethionine (AdoMet) domain and the other to three invariant cysteines in the N-terminal UPF0004 domain. We demonstrate that holo-RimO can specifically methylthiolate the aspartate residue of a 20-mer peptide derived from S12, yielding a mixture of mono- and bismethylthio derivatives. Finally, we present the 2.0 A crystal structure of the central radical AdoMet and the C-terminal TRAM (tRNA methyltransferase 2 and MiaB) domains in apo-RimO. Although the core of the open triose-phosphate isomerase (TIM) barrel of the radical AdoMet domain was conserved, RimO showed differences in domain organization compared with other radical AdoMet enzymes. The unusually acidic TRAM domain, likely to bind the basic S12 protein, is located at the distal edge of the radical AdoMet domain. The basic S12 protein substrate is likely to bind RimO through interactions with both the TRAM domain and the concave surface of the incomplete TIM barrel. These biophysical results provide a foundation for understanding the mechanism of methylthioation by radical AdoMet enzymes in the MiaB/RimO family.
Figures
Similar articles
-
Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily.Biochemistry. 2009 Oct 27;48(42):10162-74. doi: 10.1021/bi900939w. Biochemistry. 2009. PMID: 19736993 Free PMC article.
-
RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli.Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1826-31. doi: 10.1073/pnas.0708608105. Epub 2008 Feb 5. Proc Natl Acad Sci U S A. 2008. PMID: 18252828 Free PMC article.
-
Stereochemical Course of the Reaction Catalyzed by RimO, a Radical SAM Methylthiotransferase.J Am Chem Soc. 2016 Mar 9;138(9):2889-92. doi: 10.1021/jacs.5b11035. Epub 2016 Feb 25. J Am Chem Soc. 2016. PMID: 26871608
-
Structural diversity in the AdoMet radical enzyme superfamily.Biochim Biophys Acta. 2012 Nov;1824(11):1178-95. doi: 10.1016/j.bbapap.2012.04.006. Epub 2012 Apr 28. Biochim Biophys Acta. 2012. PMID: 22579873 Free PMC article. Review.
-
SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes.J Biol Chem. 2015 Feb 13;290(7):3964-71. doi: 10.1074/jbc.R114.581249. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477505 Free PMC article. Review.
Cited by
-
Characterization of LipS1 and LipS2 from Thermococcus kodakarensis: Proteins Annotated as Biotin Synthases, which Together Catalyze Formation of the Lipoyl Cofactor.ACS Bio Med Chem Au. 2022 Oct 19;2(5):509-520. doi: 10.1021/acsbiomedchemau.2c00018. Epub 2022 Jul 14. ACS Bio Med Chem Au. 2022. PMID: 36281299 Free PMC article.
-
Radical SAM enzymes in methylation and methylthiolation.Metallomics. 2012 Nov;4(11):1149-54. doi: 10.1039/c2mt20136d. Epub 2012 Sep 19. Metallomics. 2012. PMID: 22992596 Free PMC article. Review.
-
Emerging themes in radical SAM chemistry.Curr Opin Struct Biol. 2012 Dec;22(6):701-10. doi: 10.1016/j.sbi.2012.10.005. Epub 2012 Nov 8. Curr Opin Struct Biol. 2012. PMID: 23141873 Free PMC article. Review.
-
Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia.Int J Mol Sci. 2020 Feb 12;21(4):1230. doi: 10.3390/ijms21041230. Int J Mol Sci. 2020. PMID: 32059409 Free PMC article.
-
Recent advances in radical SAM enzymology: new structures and mechanisms.ACS Chem Biol. 2014 Sep 19;9(9):1929-38. doi: 10.1021/cb5004674. Epub 2014 Jul 16. ACS Chem Biol. 2014. PMID: 25009947 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

