Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms
- PMID: 20007369
- PMCID: PMC2799887
- DOI: 10.1073/pnas.0911354106
Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms
Abstract
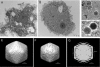
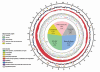
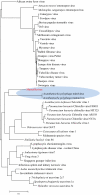
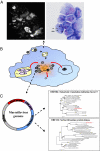
Giant viruses such as Mimivirus isolated from amoeba found in aquatic habitats show biological sophistication comparable to that of simple cellular life forms and seem to evolve by similar mechanisms, including extensive gene duplication and horizontal gene transfer (HGT), possibly in part through a viral parasite, the virophage. We report here the isolation of "Marseille" virus, a previously uncharacterized giant virus of amoeba. The virions of Marseillevirus encompass a 368-kb genome, a minimum of 49 proteins, and some messenger RNAs. Phylogenetic analysis of core genes indicates that Marseillevirus is the prototype of a family of nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes. The genome repertoire of the virus is composed of typical NCLDV core genes and genes apparently obtained from eukaryotic hosts and their parasites or symbionts, both bacterial and viral. We propose that amoebae are "melting pots" of microbial evolution where diverse forms emerge, including giant viruses with complex gene repertoires of various origins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lwoff A. The concept of virus. J Gen Microbiol. 1957;17:239–253. - PubMed
-
- Raoult D, Forterre P. Redefining viruses: Lessons from Mimivirus. Nat Rev Microbiol. 2008;6:315–319. - PubMed
-
- Van Etten JL, Meints RH. Giant viruses infecting algae. Annu Rev Microbiol. 1999;53:447–494. - PubMed
-
- La Scola B, et al. A giant virus in amoebae. Science. 2003;299:2033. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

