SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in Arabidopsis
- PMID: 20007451
- PMCID: PMC2815852
- DOI: 10.1104/pp.109.146183
SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in Arabidopsis
Abstract
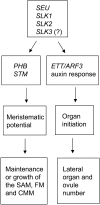
Multimeric protein complexes are required during development to regulate transcription and orchestrate cellular proliferation and differentiation. The Arabidopsis (Arabidopsis thaliana) SEUSS (SEU) gene encodes a transcriptional adaptor that shares sequence similarity with metazoan Lim domain-binding transcriptional adaptors. In Arabidopsis, SEU forms a physical complex with the LEUNIG transcriptional coregulator. This complex regulates a number of diverse developmental events, including proper specification of floral organ identity and number and the development of female reproductive tissues derived from the carpel margin meristem. In addition to SEU, there are three Arabidopsis SEUSS-LIKE (SLK) genes that encode putative transcriptional adaptors. To determine the functions of the SLK genes and to investigate the degree of functional redundancy between SEU and SLK genes, we characterized available slk mutant lines in Arabidopsis. Here, we show that mutations in any single SLK gene failed to condition an obvious morphological abnormality. However, by generating higher order mutant plants, we uncovered a degree of redundancy between the SLK genes and between SLK genes and SEU. We report a novel role for SEU and the SLK genes during embryonic development and show that the concomitant loss of both SEU and SLK2 activities conditions severe embryonic and seedling defects characterized by a loss of the shoot apical meristem. Furthermore, we demonstrate that SLK gene function is required for proper development of vital female reproductive tissues derived from the carpel margin. We propose a model that posits that SEU and SLK genes support organ development from meristematic regions through two different pathways: one that facilitates auxin response and thus organ initiation and a second that sustains meristematic potential through the maintenance of SHOOTMERISTEM-LESS and PHABULOSA expression.
Figures




References
-
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H (1996) Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384 270–272 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

