Frame-disrupting mutations elicit pre-mRNA accumulation independently of frame disruption
- PMID: 20007599
- PMCID: PMC2836556
- DOI: 10.1093/nar/gkp1115
Frame-disrupting mutations elicit pre-mRNA accumulation independently of frame disruption
Abstract
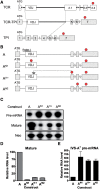
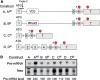
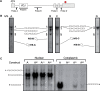
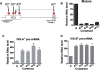
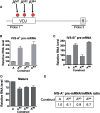
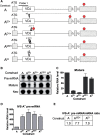
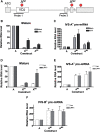
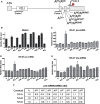
The T-cell receptor (TCR) and immunoglobulin (Ig) genes are unique among vertebrate genes in that they undergo programmed rearrangement, a process that allows them to generate an enormous array of receptors with different antigen specificities. While crucial for immune function, this rearrangement mechanism is highly error prone, often generating frameshift or nonsense mutations that render the rearranged TCR and Ig genes defective. Such frame-disrupting mutations have been reported to increase the level of TCRbeta and Igmicro pre-mRNA, suggesting the hypothesis that RNA processing is blocked when frame disruption is sensed. Using a chimeric gene that contains TCRbeta sequences conferring this upregulatory response, we provide evidence that pre-mRNA upregulation is neither frame- nor translation-dependent; instead, several lines of evidence suggested that it is the result of disrupted cis elements necessary for efficient RNA splicing. In particular, we identify the rearranging VDJ(beta) exon as being uniquely densely packed with exonic-splicing enhancers (ESEs), rendering this exon hypersensitive to mutational disruption. As the chimeric gene that we developed for these studies generates unusually stable nuclear pre-mRNAs that accumulate when challenged with ESE mutations, we suggest it can be used as a sensitive in vivo system to identify and characterize ESEs.
Figures

References
-
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. - PubMed
-
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. - PubMed
-
- Bruce SR, Wilkinson MF. Nonsense-Mediated Decay: A Surveillance Pathway that Detects Faulty TCR and BCR Transcripts. Trivandrum, India: Research Signpost; 2003.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

