Involvement of Vts1, a structure-specific RNA-binding protein, in Okazaki fragment processing in yeast
- PMID: 20007605
- PMCID: PMC2836565
- DOI: 10.1093/nar/gkp1135
Involvement of Vts1, a structure-specific RNA-binding protein, in Okazaki fragment processing in yeast
Abstract
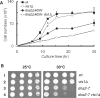
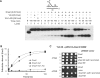
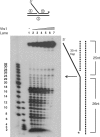
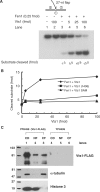
The non-essential VTS1 gene of Saccharomyces cerevisiae is highly conserved in eukaryotes and encodes a sequence- and structure-specific RNA-binding protein. The Vts1 protein has been implicated in post-transcriptional regulation of a specific set of mRNAs that contains its-binding site at their 3'-untranslated region. In this study, we identified VTS1 as a multi-copy suppressor of dna2-K1080E, a lethal mutant allele of DNA2 that lacks DNA helicase activity. The suppression was allele-specific, since overexpression of Vts1 did not suppress the temperature-dependent growth defects of dna2Delta405N devoid of the N-terminal 405-amino-acid residues. Purified recombinant Vts1 stimulated the endonuclease activity of wild-type Dna2, but not the endonuclease activity of Dna2Delta405N, indicating that the activation requires the N-terminal domain of Dna2. Stimulation of Dna2 endonuclease activity by Vts1 appeared to be the direct cause of suppression, since the multi-copy expression of Dna2-K1080E suppressed the lethality observed with its single-copy expression. We found that vts1Delta dna2Delta405N and vts1Deltadna2-7 double mutant cells displayed synergistic growth defects, in support of a functional interaction between two genes. Our results provide both in vivo and in vitro evidence that Vts1 is involved in lagging strand synthesis by modulating the Dna2 endonuclease activity that plays an essential role in Okazaki fragment processing.
Figures





Similar articles
-
Rad52/Rad59-dependent recombination as a means to rectify faulty Okazaki fragment processing.J Biol Chem. 2014 May 23;289(21):15064-79. doi: 10.1074/jbc.M114.548388. Epub 2014 Apr 7. J Biol Chem. 2014. PMID: 24711454 Free PMC article.
-
The trans-autostimulatory activity of Rad27 suppresses dna2 defects in Okazaki fragment processing.J Biol Chem. 2012 Mar 16;287(12):8675-87. doi: 10.1074/jbc.M111.326470. Epub 2012 Jan 9. J Biol Chem. 2012. PMID: 22235122 Free PMC article.
-
In vivo and in vitro studies of Mgs1 suggest a link between genome instability and Okazaki fragment processing.Nucleic Acids Res. 2005 Oct 26;33(19):6137-50. doi: 10.1093/nar/gki900. Print 2005. Nucleic Acids Res. 2005. PMID: 16251400 Free PMC article.
-
Yet another job for Dna2: Checkpoint activation.DNA Repair (Amst). 2015 Aug;32:17-23. doi: 10.1016/j.dnarep.2015.04.009. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956863 Free PMC article. Review.
-
Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes.Crit Rev Biochem Mol Biol. 2010 Apr;45(2):71-96. doi: 10.3109/10409230903578593. Crit Rev Biochem Mol Biol. 2010. PMID: 20131965 Review.
Cited by
-
Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast.Mol Biol Cell. 2011 May 15;22(10):1648-63. doi: 10.1091/mbc.E10-11-0878. Epub 2011 Mar 25. Mol Biol Cell. 2011. PMID: 21441304 Free PMC article.
-
Rad52/Rad59-dependent recombination as a means to rectify faulty Okazaki fragment processing.J Biol Chem. 2014 May 23;289(21):15064-79. doi: 10.1074/jbc.M114.548388. Epub 2014 Apr 7. J Biol Chem. 2014. PMID: 24711454 Free PMC article.
-
Saccharomyces cerevisiae Cmr1 protein preferentially binds to UV-damaged DNA in vitro.J Microbiol. 2012 Feb;50(1):112-8. doi: 10.1007/s12275-012-1597-4. Epub 2012 Feb 27. J Microbiol. 2012. PMID: 22367945
-
GSK-3β Homolog Rim11 and the Histone Deacetylase Complex Ume6-Sin3-Rpd3 Are Involved in Replication Stress Response Caused by Defects in Dna2.Genetics. 2017 Jun;206(2):829-842. doi: 10.1534/genetics.116.198671. Epub 2017 May 3. Genetics. 2017. PMID: 28468907 Free PMC article.
-
The trans-autostimulatory activity of Rad27 suppresses dna2 defects in Okazaki fragment processing.J Biol Chem. 2012 Mar 16;287(12):8675-87. doi: 10.1074/jbc.M111.326470. Epub 2012 Jan 9. J Biol Chem. 2012. PMID: 22235122 Free PMC article.
References
-
- MacNeill SA. DNA replication: partners in the Okazaki two-step. Curr. Biol. 2001;11:R842–R844. - PubMed
-
- Hübscher U, Seo YS. Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells. 2001;12:149–157. - PubMed
-
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. - PubMed
-
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. - PubMed
-
- Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 2006;106:453–473. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

