Alterations of the Notch pathway in lung cancer
- PMID: 20007775
- PMCID: PMC2799768
- DOI: 10.1073/pnas.0907781106
Alterations of the Notch pathway in lung cancer
Abstract
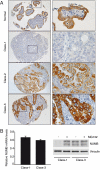
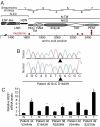
Notch signaling regulates cell specification and homeostasis of stem cell compartments, and it is counteracted by the cell fate determinant Numb. Both Numb and Notch have been implicated in human tumors. Here, we show that Notch signaling is altered in approximately one third of non-small-cell lung carcinomas (NSCLCs), which are the leading cause of cancer-related deaths: in approximately 30% of NSCLCs, loss of Numb expression leads to increased Notch activity, while in a smaller fraction of cases (around 10%), gain-of-function mutations of the NOTCH-1 gene are present. Activation of Notch correlates with poor clinical outcomes in NSCLC patients without TP53 mutations. Finally, primary epithelial cell cultures, derived from NSCLC harboring constitutive activation of the Notch pathway, are selectively killed by inhibitors of Notch (gamma-secretase inhibitors), showing that the proliferative advantage of these tumors is dependent upon Notch signaling. Our results show that the deregulation of the Notch pathway is a relatively frequent event in NSCLCs and suggest that it might represent a possible target for molecular therapies in these tumors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. - PubMed
-
- Gonczy P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. - PubMed
-
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. - PubMed
-
- Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

