Usefulness of peptide nucleic acid (PNA)-clamp smart amplification process version 2 (SmartAmp2) for clinical diagnosis of KRAS codon 12 mutations in lung adenocarcinoma: comparison of PNA-clamp SmartAmp2 and PCR-related methods
- PMID: 20007840
- PMCID: PMC2797726
- DOI: 10.2353/jmoldx.2010.090081
Usefulness of peptide nucleic acid (PNA)-clamp smart amplification process version 2 (SmartAmp2) for clinical diagnosis of KRAS codon 12 mutations in lung adenocarcinoma: comparison of PNA-clamp SmartAmp2 and PCR-related methods
Abstract
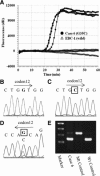
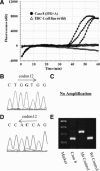
KRAS is an oncogene that can be activated by mutations. Patients with non-small cell lung cancer who have KRAS mutations do not respond to tyrosine kinase inhibitors; therefore, accurate detection of KRAS mutations is important for deciding therapeutic strategies. Although sequencing-related techniques have been frequently used, they are usually too complex, have low sensitivity, and are time-consuming for routine screening in clinical situations. We evaluated peptide nucleic acid (PNA)-clamp smart amplification process version 2 (SmartAmp2) as a detection method for KRAS codon 12 mutations in patient specimens compared with traditional sequencing and polymerase chain reaction-related methods. Among 172 lung adenocarcinoma samples, direct sequencing, enzyme-enriched sequencing, and PNA-enriched sequencing showed that 16 (9.3%), 26 (15.7%), and 28 (16.3%) tumors, respectively, contained KRAS mutations in codon 12. Using PNA-clamp SmartAmp2, we could identify 31 (18.0%) tumors that had KRAS mutations in codon 12 within 60 minutes, three of which were undetected by polymerase chain reaction-related methods. On the other hand, we examined 30 nonmalignant peripheral lung tissue specimens and found no mutations in any of the samples using PNA-clamp SmartAmp2. In this study, we confirmed that PNA-clamp SmartAmp2 has high sensitivity and accuracy and is suitable for the clinical diagnosis of KRAS codon 12 mutations.
Figures
References
-
- Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. - PubMed
-
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. - PubMed
-
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain't over ‘til it's over’. Trends Cell Biol. 2000;10:147–154. - PubMed
-
- Vojtek AB, Cooper JA. Rho family members: activators of MAP kinase cascades. Cell. 1995;82:527–529. - PubMed
Uncited reference
-
- Ramos FJ, Macarulla T, Capdevila J, Elez E, Tabernero J. Understanding the predictive role of K-ras for epidermal growth factor receptor-targeted therapies in colorectal cancer. Clin Colorectal Cancer. 2008;7:S52–S57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

