KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer
- PMID: 20007845
- PMCID: PMC2797717
- DOI: 10.2353/jmoldx.2010.080131
KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer
Abstract
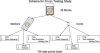
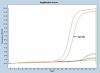
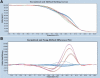
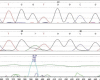
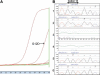
Treatment of colon carcinoma with the anti-epidermal growth factor receptor antibody Cetuximab is reported to be ineffective in KRAS-mutant tumors. Mutation testing techniques have therefore become an urgent concern. We have compared three methods for detecting KRAS mutations in 59 cases of colon carcinoma: 1) high resolution melting, 2) the amplification refractory mutation system using a bifunctional self-probing primer (ARMS/Scorpion, ARMS/S), and 3) direct sequencing. We also evaluated the effects of the methods of sectioning and coring of paraffin blocks to obtain tumor DNA on assay sensitivity and specificity. The most sensitive and specific combination of block sampling and mutational analysis was ARMS/S performed on DNA derived from 1-mm paraffin cores. This combination of tissue sampling and testing method detected KRAS mutations in 46% of colon tumors. Four samples were positive by ARMS/S, but initially negative by direct sequencing. Cloned DNA samples were retested by direct sequencing, and in all four cases KRAS mutations were identified in the DNA. In six cases, high resolution melting abnormalities could not be confirmed as specific mutations either by ARMS/S or direct sequencing. We conclude that coring of the paraffin blocks and testing by ARMS/S is a sensitive, specific, and efficient method for KRAS testing.
Figures
References
-
- Shih C, Padhy LC, Murray M, Weinberg RA. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–264. - PubMed
-
- Slebos RJ, Rodenhuis S. The molecular genetics of human lung cancer. Eur Respir J. 1989;2:461–469. - PubMed
-
- Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

