High-throughput identification and quantification of Candida species using high resolution derivative melt analysis of panfungal amplicons
- PMID: 20007848
- PMCID: PMC2797723
- DOI: 10.2353/jmoldx.2010.090085
High-throughput identification and quantification of Candida species using high resolution derivative melt analysis of panfungal amplicons
Abstract
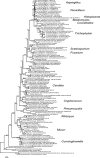
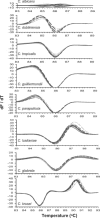
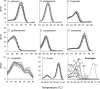
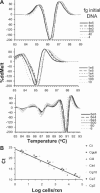
Fungal infections pose unique challenges to molecular diagnostics; fungal molecular diagnostics consequently lags behind bacterial and viral counterparts. Nevertheless, fungal infections are often life-threatening, and early detection and identification of species is crucial to successful intervention. A high throughput PCR-based method is needed that is independent of culture, is sensitive to the level of one fungal cell per milliliter of blood or other tissue types, and is capable of detecting species and resistance mutations. We introduce the use of high resolution melt analysis, in combination with more sensitive, inclusive, and appropriately positioned panfungal primers, to address these needs. PCR-based amplification of the variable internal transcribed regions of the rDNA genes generates an amplicon whose sequence melts with a shape that is characteristic and therefore diagnostic of the species. Simple analysis of the differences between test and reference melt curves generates a single number that calls the species. Early indications suggest that high resolution melt analysis can distinguish all eight major species of Candida of clinical significance without interference from excess human DNA. Candida species, including mixed and novel species, can be identified directly in vaginal samples. This tool can potentially detect, count, and identify fungi in hundreds of samples per day without further manipulation, costs, or delays, offering a major step forward in fungal molecular diagnostics.
Figures

References
-
- White PL, Barton R, Guiver M, Linton CJ, Wilson S, Smith M, Gomez BL, Carr MJ, Kimmitt PT, Seaton S, Rajakumar K, Holyoake T, Kibbler CC, Johnson E, Hobson RP, Jones B, Barnes RA. A consensus on fungal polymerase chain reaction diagnosis? a United Kingdom-Ireland evaluation of polymerase chain reaction methods for detection of systemic fungal infections. J Mol Diagn. 2006;8:376–384. - PMC - PubMed
-
- Baskova L, Landlinger C, Preuner S, Lion T. The Pan-AC assay: a single-reaction real-time PCR test for quantitative detection of a broad range of Aspergillus and Candida species. J Med Microbiol. 2007;56:1167–1173. - PubMed
-
- De Marco D, Perotti M, Ossi CM, Burioni R, Clementi M, Mancini N. Development and validation of a molecular method for the diagnosis of medically important fungal infections. New Microbiol. 2007;30:308–312. - PubMed
-
- Dial SM. Fungal diagnostics: current techniques and future trends. Vet Clinics N Am. 2007;37:373–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

