Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing
- PMID: 20007951
- PMCID: PMC2824209
- DOI: 10.1074/jbc.M109.055996
Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing
Abstract
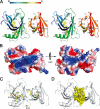
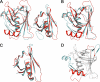
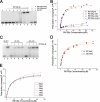
Rtt106p is a Saccharomyces cerevisiae histone chaperone with roles in heterochromatin silencing and nucleosome assembly. The molecular mechanism by which Rtt106p engages in chromatin dynamics remains unclear. Here, we report the 2.5 A crystal structure of the core domain of Rtt106p, which adopts an unusual "double pleckstrin homology" domain architecture that represents a novel structural mode for histone chaperones. A histone H3-H4-binding region and a novel double-stranded DNA-binding region have been identified. Mutagenesis studies reveal that the histone and DNA binding activities of Rtt106p are involved in Sir protein-mediated heterochromatin formation. Our results uncover the structural basis of the diverse functions of Rtt106p and provide new insights into its cellular roles.
Figures




Similar articles
-
A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing.EMBO J. 2007 May 2;26(9):2274-83. doi: 10.1038/sj.emboj.7601670. Epub 2007 Apr 5. EMBO J. 2007. PMID: 17410207 Free PMC article.
-
A region of the nucleosome required for multiple types of transcriptional silencing in Saccharomyces cerevisiae.Genetics. 2011 Jul;188(3):535-48. doi: 10.1534/genetics.111.129197. Epub 2011 May 5. Genetics. 2011. PMID: 21546544 Free PMC article.
-
Novel functional residues in the core domain of histone H2B regulate yeast gene expression and silencing and affect the response to DNA damage.Mol Cell Biol. 2010 Jul;30(14):3503-18. doi: 10.1128/MCB.00290-10. Epub 2010 May 17. Mol Cell Biol. 2010. PMID: 20479120 Free PMC article.
-
SIR proteins and the assembly of silent chromatin in budding yeast.Annu Rev Genet. 2013;47:275-306. doi: 10.1146/annurev-genet-021313-173730. Epub 2013 Sep 4. Annu Rev Genet. 2013. PMID: 24016189 Review.
-
The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways.Chromosoma. 2007 Apr;116(2):79-93. doi: 10.1007/s00412-006-0087-z. Epub 2006 Dec 19. Chromosoma. 2007. PMID: 17180700 Review.
Cited by
-
Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.Proc Natl Acad Sci U S A. 2012 Jan 17;109(3):E144-53. doi: 10.1073/pnas.1119095109. Epub 2011 Dec 23. Proc Natl Acad Sci U S A. 2012. PMID: 22198837 Free PMC article.
-
Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression.Mol Cell Biol. 2012 Nov;32(21):4337-49. doi: 10.1128/MCB.00871-12. Epub 2012 Aug 20. Mol Cell Biol. 2012. PMID: 22907759 Free PMC article.
-
Modulation of Gene Silencing by Cdc7p via H4 K16 Acetylation and Phosphorylation of Chromatin Assembly Factor CAF-1 in Saccharomyces cerevisiae.Genetics. 2019 Apr;211(4):1219-1237. doi: 10.1534/genetics.118.301858. Epub 2019 Feb 6. Genetics. 2019. PMID: 30728156 Free PMC article.
-
Redundant Functions for Nap1 and Chz1 in H2A.Z Deposition.Sci Rep. 2017 Sep 7;7(1):10791. doi: 10.1038/s41598-017-11003-8. Sci Rep. 2017. PMID: 28883625 Free PMC article.
-
Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1.Nucleic Acids Res. 2018 Nov 2;46(19):9907-9917. doi: 10.1093/nar/gky823. Nucleic Acids Res. 2018. PMID: 30239791 Free PMC article. Review.
References
-
- Imbeault D., Gamar L., Rufiange A., Paquet E., Nourani A. (2008) J. Biol. Chem. 283, 27350–27354 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

