Pattern recognition receptors require N-glycosylation to mediate plant immunity
- PMID: 20007973
- PMCID: PMC2836068
- DOI: 10.1074/jbc.M109.063073
Pattern recognition receptors require N-glycosylation to mediate plant immunity
Abstract
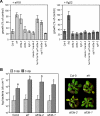
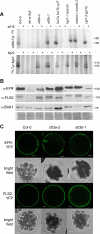
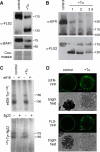
N-Glycans attached to the ectodomains of plasma membrane pattern recognition receptors constitute likely initial contact sites between plant cells and invading pathogens. To assess the role of N-glycans in receptor-mediated immune responses, we investigated the functionality of Arabidopsis receptor kinases EFR and FLS2, sensing bacterial translation elongation factor Tu (elf18) and flagellin (flg22), respectively, in N-glycosylation mutants. As revealed by binding and responses to elf18 or flg22, both receptors tolerated immature N-glycans induced by mutations in various Golgi modification steps. EFR was specifically impaired by loss-of-function mutations in STT3A, a subunit of the endoplasmic reticulum resident oligosaccharyltransferase complex. FLS2 tolerated mild underglycosylation occurring in stt3a but was sensitive to severe underglycosylation induced by tunicamycin treatment. EFR accumulation was significantly reduced when synthesized without N-glycans but to lesser extent when underglycosylated in stt3a or mutated in single amino acid positions. Interestingly, EFR(N143Q) lacking a single conserved N-glycosylation site from the EFR ectodomain accumulated to reduced levels and lost the ability to bind its ligand and to mediate elf18-elicited oxidative burst. However, EFR-YFP protein localization and peptide:N-glycosidase F digestion assays support that both EFR produced in stt3a and EFR(N143Q) in wild type cells correctly targeted to the plasma membrane via the Golgi apparatus. These results indicate that a single N-glycan plays a critical role for receptor abundance and ligand recognition during plant-pathogen interactions at the cell surface.
Figures

References
-
- Elbein A. D. (1987) Annu. Rev. Biochem. 56, 497–534 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

