Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968
- PMID: 20008069
- PMCID: PMC2812971
- DOI: 10.1128/JB.01272-09
Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968
Abstract
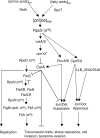
Legionella longbeachae causes most cases of legionellosis in Australia and may be underreported worldwide due to the lack of L. longbeachae-specific diagnostic tests. L. longbeachae displays distinctive differences in intracellular trafficking, caspase 1 activation, and infection in mouse models compared to Legionella pneumophila, yet these two species have indistinguishable clinical presentations in humans. Unlike other legionellae, which inhabit freshwater systems, L. longbeachae is found predominantly in moist soil. In this study, we sequenced and annotated the genome of an L. longbeachae clinical isolate from Oregon, isolate D-4968, and compared it to the previously published genomes of L. pneumophila. The results revealed that the D-4968 genome is larger than the L. pneumophila genome and has a gene order that is different from that of the L. pneumophila genome. Genes encoding structural components of type II, type IV Lvh, and type IV Icm/Dot secretion systems are conserved. In contrast, only 42/140 homologs of genes encoding L. pneumophila Icm/Dot substrates have been found in the D-4968 genome. L. longbeachae encodes numerous proteins with eukaryotic motifs and eukaryote-like proteins unique to this species, including 16 ankyrin repeat-containing proteins and a novel U-box protein. We predict that these proteins are secreted by the L. longbeachae Icm/Dot secretion system. In contrast to the L. pneumophila genome, the L. longbeachae D-4968 genome does not contain flagellar biosynthesis genes, yet it contains a chemotaxis operon. The lack of a flagellum explains the failure of L. longbeachae to activate caspase 1 and trigger pyroptosis in murine macrophages. These unique features of L. longbeachae may reflect adaptation of this species to life in soil.
Figures


References
-
- Anonymous. 2000. Legionnaires' disease associated with potting soil—California, Oregon, and Washington, May-June 2000. MMWR Morb. Mortal. Wkly. Rep. 49:777-778. - PubMed
-
- Asare, R., and Y. Abu Kwaik. 2007. Early trafficking and intracellular replication of Legionella longbeachae within an ER-derived late endosome-like phagosome. Cell. Microbiol. 9:1571-1587. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

