Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae
- PMID: 20008079
- PMCID: PMC2837980
- DOI: 10.1128/EC.00213-09
Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae
Abstract
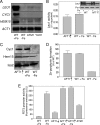
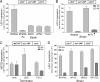
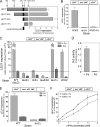
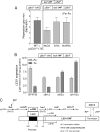
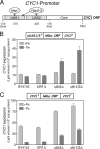
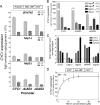
Budding yeast (Saccharomyces cerevisiae) responds to iron deprivation both by Aft1-Aft2-dependent transcriptional activation of genes involved in cellular iron uptake and by Cth1-Cth2-specific degradation of certain mRNAs coding for iron-dependent biosynthetic components. Here, we provide evidence for a novel principle of iron-responsive gene expression. This regulatory mechanism is based on the modulation of transcription through the iron-dependent variation of levels of regulatory metabolites. As an example, the LEU1 gene of branched-chain amino acid biosynthesis is downregulated under iron-limiting conditions through depletion of the metabolic intermediate alpha-isopropylmalate, which functions as a key transcriptional coactivator of the Leu3 transcription factor. Synthesis of alpha-isopropylmalate involves the iron-sulfur protein Ilv3, which is inactivated under iron deficiency. As another example, decreased mRNA levels of the cytochrome c-encoding CYC1 gene under iron-limiting conditions involve heme-dependent transcriptional regulation via the Hap1 transcription factor. Synthesis of the iron-containing heme is directly correlated with iron availability. Thus, the iron-responsive expression of genes that are downregulated under iron-limiting conditions is conferred by two independent regulatory mechanisms: transcriptional regulation through iron-responsive metabolites and posttranscriptional mRNA degradation. Only the combination of the two processes provides a quantitative description of the response to iron deprivation in yeast.
Figures
Similar articles
-
Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation.Mol Cell Biol. 2013 Jun;33(11):2178-87. doi: 10.1128/MCB.01458-12. Epub 2013 Mar 25. Mol Cell Biol. 2013. PMID: 23530061 Free PMC article.
-
Cisplatin upregulates Saccharomyces cerevisiae genes involved in iron homeostasis through activation of the iron insufficiency-responsive transcription factor Aft1.J Cell Physiol. 2007 Feb;210(2):378-84. doi: 10.1002/jcp.20845. J Cell Physiol. 2007. PMID: 17096368
-
Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1.Mol Cell Biol. 2005 Aug;25(15):6760-71. doi: 10.1128/MCB.25.15.6760-6771.2005. Mol Cell Biol. 2005. PMID: 16024809 Free PMC article.
-
Post-transcriptional regulation of iron homeostasis in Saccharomyces cerevisiae.Int J Mol Sci. 2013 Jul 30;14(8):15785-809. doi: 10.3390/ijms140815785. Int J Mol Sci. 2013. PMID: 23903042 Free PMC article. Review.
-
Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae.Genes (Basel). 2020 Jul 15;11(7):795. doi: 10.3390/genes11070795. Genes (Basel). 2020. PMID: 32679672 Free PMC article. Review.
Cited by
-
Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation.Mol Cell Biol. 2013 Jun;33(11):2178-87. doi: 10.1128/MCB.01458-12. Epub 2013 Mar 25. Mol Cell Biol. 2013. PMID: 23530061 Free PMC article.
-
Regulation of ribonucleotide reductase in response to iron deficiency.Mol Cell. 2011 Dec 9;44(5):759-69. doi: 10.1016/j.molcel.2011.09.021. Mol Cell. 2011. PMID: 22152479 Free PMC article.
-
Iron Deficiency and Recovery in Yeast: A Quantitative Proteomics Approach.J Proteome Res. 2021 May 7;20(5):2751-2761. doi: 10.1021/acs.jproteome.1c00035. Epub 2021 Apr 2. J Proteome Res. 2021. PMID: 33797912 Free PMC article.
-
Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation.Adv Microb Physiol. 2012;60:91-210. doi: 10.1016/B978-0-12-398264-3.00002-4. Adv Microb Physiol. 2012. PMID: 22633059 Free PMC article. Review.
-
HAL2 overexpression induces iron acquisition in bdf1Δ cells and enhances their salt resistance.Curr Genet. 2017 May;63(2):229-239. doi: 10.1007/s00294-016-0628-9. Epub 2016 Jul 8. Curr Genet. 2017. PMID: 27387517
References
-
- Becerra M., Lombardia-Ferreira L. J., Hauser N. C., Hoheisel J. D., Tizon B., Cerdan M. E. 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43:545–555 - PubMed
-
- Belli G., Molina M. M., Garcia-Martinez J., Perez-Ortin J. E., Herrero E. 2004. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279:12386–12395 - PubMed
-
- Camadro J. M., Labbe P. 1988. Purification and properties of ferrochelatase from the yeast Saccharomyces cerevisiae. Evidence for a precursor form of the protein. J. Biol. Chem. 263:11675–11682 - PubMed
-
- Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., Kaplan J. 2004. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279:29513–29518 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

