Quantitation of cellular deoxynucleoside triphosphates
- PMID: 20008099
- PMCID: PMC2847218
- DOI: 10.1093/nar/gkp1141
Quantitation of cellular deoxynucleoside triphosphates
Abstract
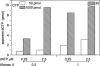
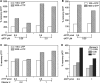
Eukaryotic cells contain a delicate balance of minute amounts of the four deoxyribonucleoside triphosphates (dNTPs), sufficient only for a few minutes of DNA replication. Both a deficiency and a surplus of a single dNTP may result in increased mutation rates, faulty DNA repair or mitochondrial DNA depletion. dNTPs are usually quantified by an enzymatic assay in which incorporation of radioactive dATP (or radioactive dTTP in the assay for dATP) into specific synthetic oligonucleotides by a DNA polymerase is proportional to the concentration of the unknown dNTP. We find that the commonly used Klenow DNA polymerase may substitute the corresponding ribonucleotide for the unknown dNTP leading in some instances to a large overestimation of dNTPs. We now describe assay conditions for each dNTP that avoid ribonucleotide incorporation. For the dTTP and dATP assays it suffices to minimize the concentrations of the Klenow enzyme and of labeled dATP (or dTTP); for dCTP and dGTP we had to replace the Klenow enzyme with either the Taq DNA polymerase or Thermo Sequenase. We suggest that in some earlier reports ribonucleotide incorporation may have caused too high values for dGTP and dCTP.
Figures

Similar articles
-
Quantitation of deoxynucleoside triphosphates by click reactions.Sci Rep. 2020 Jan 17;10(1):611. doi: 10.1038/s41598-020-57463-3. Sci Rep. 2020. PMID: 31953472 Free PMC article.
-
An improved method for the quantitative determination of deoxyribonucleoside triphosphates in cell extracts.Anal Biochem. 1984 May 15;139(1):35-41. doi: 10.1016/0003-2697(84)90386-5. Anal Biochem. 1984. PMID: 6331228
-
Deoxyribonucleoside triphosphate pools in human diploid fibroblasts and their modulation by hydroxyurea and deoxynucleosides.Biochem Pharmacol. 1984 May 1;33(9):1515-8. doi: 10.1016/0006-2952(84)90421-0. Biochem Pharmacol. 1984. PMID: 6732868
-
Changes in deoxynucleoside triphosphate pools induced by inhibitors and modulators of ribonucleotide reductase.Pharmacol Ther. 1985;30(1):31-42. doi: 10.1016/0163-7258(85)90046-4. Pharmacol Ther. 1985. PMID: 3915820 Review.
-
A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells.Nucleosides Nucleotides Nucleic Acids. 2011 May;30(5):317-39. doi: 10.1080/15257770.2011.586955. Nucleosides Nucleotides Nucleic Acids. 2011. PMID: 21774628 Free PMC article. Review.
Cited by
-
Quantitative Analysis of Nucleoside Triphosphate Pools in Mouse Muscle Using Hydrophilic Interaction Liquid Chromatography Coupled with Tandem Mass Spectrometry Detection.Methods Mol Biol. 2023;2615:267-280. doi: 10.1007/978-1-0716-2922-2_19. Methods Mol Biol. 2023. PMID: 36807798
-
Simultaneous isotope dilution quantification and metabolic tracing of deoxyribonucleotides by liquid chromatography high resolution mass spectrometry.Anal Biochem. 2019 Mar 1;568:65-72. doi: 10.1016/j.ab.2018.12.023. Epub 2018 Dec 31. Anal Biochem. 2019. PMID: 30605633 Free PMC article.
-
Deoxynucleoside stress exacerbates the phenotype of a mouse model of mitochondrial neurogastrointestinal encephalopathy.Brain. 2014 May;137(Pt 5):1337-49. doi: 10.1093/brain/awu068. Epub 2014 Apr 10. Brain. 2014. PMID: 24727567 Free PMC article.
-
Deoxypyrimidine monophosphate bypass therapy for thymidine kinase 2 deficiency.EMBO Mol Med. 2014 Aug;6(8):1016-27. doi: 10.15252/emmm.201404092. EMBO Mol Med. 2014. PMID: 24968719 Free PMC article.
-
HIV-1 Reverse Transcriptase-Based Assay to Determine Cellular dNTP Concentrations.Methods Mol Biol. 2016;1354:61-70. doi: 10.1007/978-1-4939-3046-3_5. Methods Mol Biol. 2016. PMID: 26714705 Free PMC article.
References
-
- Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp.Cell Res. 1989;181:305–316. - PubMed
-
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis (1988) Annu. Rev. Biochem. 1988;57:349–374. - PubMed
-
- Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, MacIntosh EM, Reidy IA. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. - PubMed
-
- Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial intergenomic signaling. Gene. 2005;354:162–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

