The role of NOTCH1 signaling in T-ALL
- PMID: 20008221
- PMCID: PMC2847371
- DOI: 10.1182/asheducation-2009.1.353
The role of NOTCH1 signaling in T-ALL
Abstract
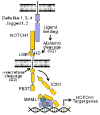
The identification of activating mutations in NOTCH1 in over 50% of T-cell acute lymphoblastic leukemias (T-ALL) has generated major interest in the elucidation of the mechanisms of transformation downstream of oncogenic NOTCH and in the targeting of the NOTCH signaling pathway in this disease. Small molecule gamma-secretase inhibitors (GSIs) block NOTCH1 signaling in T-ALL lymphoblasts, yet the clinical development of GSIs has been held back by the development of gastrointestinal toxicity and their weak antileukemic effects against human T-ALL. However, new therapeutic strategies aiming to optimize the use of anti-NOTCH1 therapies for T-ALL, including combination therapies with molecularly targeted drugs and glucocorticoids, have started to emerge as a result of improved understanding of the molecular mechanisms that mediate the effects of GSIs in leukemic cells and the intestinal epithelium. This review focuses on the molecular basis of NOTCH1-induced transformation, the mechanisms of action of oncogenic NOTCH1 and clinical significance of NOTCH1 mutations in T-ALL.
Figures

References
-
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. - PubMed
-
- Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. - PubMed
-
- Palomero T, Barnes KC, Real PJ, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. - PubMed
-
- Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

