Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia
- PMID: 20008299
- PMCID: PMC2837338
- DOI: 10.1182/blood-2009-06-226340
Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia
Abstract
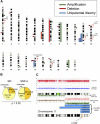
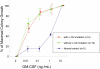
Juvenile myelomonocytic leukemia (JMML) is a rare pediatric myeloid neoplasm characterized by excessive proliferation of myelomonocytic cells. When we investigated the presence of recurrent molecular lesions in a cohort of 49 children with JMML, neurofibromatosis phenotype (and thereby NF1 mutation) was present in 2 patients (4%), whereas previously described PTPN11, NRAS, and KRAS mutations were found in 53%, 4%, and 2% of cases, respectively. Consequently, a significant proportion of JMML patients without identifiable pathogenesis prompted our search for other molecular defects. When we applied single nucleotide polymorphism arrays to JMML patients, somatic uniparental disomy 11q was detected in 4 of 49 patients; all of these cases harbored RING finger domain c-Cbl mutations. In total, c-Cbl mutations were detected in 5 (10%) of 49 patients. No mutations were identified in Cbl-b and TET2. c-Cbl and RAS pathway mutations were mutually exclusive. Comparison of clinical phenotypes showed earlier presentation and lower hemoglobin F levels in patients with c-Cbl mutations. Our results indicate that mutations in c-Cbl may represent key molecular lesions in JMML patients without RAS/PTPN11 lesions, suggesting analogous pathogenesis to those observed in chronic myelomonocytic leukemia (CMML) patients.
Figures

References
-
- Niemeyer CM, Arico M, Basso G, et al. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases: European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood. 1997;89(10):3534–3543. - PubMed
-
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77(5):925–929. - PubMed
-
- Emanuel PD. Juvenile myelomonocytic leukemia. Curr Hematol Rep. 2004;3(3):203–209. - PubMed
-
- Flotho C, Valcamonica S, Mach-Pascual S, et al. RAS mutations and clonality analysis in children with juvenile myelomonocytic leukemia (JMML). Leukemia. 1999;13(1):32–37. - PubMed
-
- Tartaglia M, Niemeyer CM, Fragale A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34(2):148–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

