Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation
- PMID: 20008320
- PMCID: PMC2824207
- DOI: 10.1074/jbc.M109.050732
Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation
Abstract
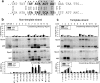
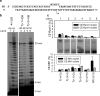
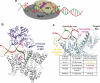
The catalytic subunit of the mitochondrial (mt) RNA polymerase (RNAP) is highly homologous to the bacteriophage T7/T3 RNAP. Unlike the phage RNAP, however, the mtRNAP relies on accessory proteins to initiate promoter-specific transcription. Rpo41, the catalytic subunit of the Saccharomyces cerevisiae mtRNAP, requires Mtf1 for opening the duplex promoter. To elucidate the role of Mtf1 in promoter-specific DNA opening, we have mapped the structural organization of the mtRNAP using site-specific protein-DNA photo-cross-linking studies. Both Mtf1 and Rpo41 cross-linked to distinct sites on the promoter DNA, but the dominant cross-links were those of the Mtf1, which indicates a direct role of Mtf1 in promoter-specific binding and initiation. Strikingly, Mtf1 cross-linked with a high efficiency to the melted region of the promoter DNA, based on which we suggest that Mtf1 facilitates DNA melting by trapping the non-template strand in the unwound conformation. Additional strong cross-links of the Mtf1 were observed with the -8 to -10 base-paired region of the promoter. The cross-linking results were incorporated into a structural model of the mtRNAP-DNA, created from a homology model of the C-terminal domain of Rpo41 and the available structure of Mtf1. The promoter DNA is sandwiched between Mtf1 and Rpo41 in the structural model, and Mtf1 closely associates mainly with one face of the promoter across the entire nona-nucleotide consensus sequence. Overall, the studies reveal that in many ways the role of Mtf1 is analogous to the transcription factors of the multisubunit RNAPs, which provides an intriguing link between single- and multisubunit RNAPs.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

