Genome-wide scans for footprints of natural selection
- PMID: 20008396
- PMCID: PMC2842710
- DOI: 10.1098/rstb.2009.0219
Genome-wide scans for footprints of natural selection
Abstract
Detecting recent selected 'genomic footprints' applies directly to the discovery of disease genes and in the imputation of the formative events that molded modern population genetic structure. The imprints of historic selection/adaptation episodes left in human and animal genomes allow one to interpret modern and ancestral gene origins and modifications. Current approaches to reveal selected regions applied in genome-wide selection scans (GWSSs) fall into eight principal categories: (I) phylogenetic footprinting, (II) detecting increased rates of functional mutations, (III) evaluating divergence versus polymorphism, (IV) detecting extended segments of linkage disequilibrium, (V) evaluating local reduction in genetic variation, (VI) detecting changes in the shape of the frequency distribution (spectrum) of genetic variation, (VII) assessing differentiating between populations (F(ST)), and (VIII) detecting excess or decrease in admixture contribution from one population. Here, we review and compare these approaches using available human genome-wide datasets to provide independent verification (or not) of regions found by different methods and using different populations. The lessons learned from GWSSs will be applied to identify genome signatures of historic selective pressures on genes and gene regions in other species with emerging genome sequences. This would offer considerable potential for genome annotation in functional, developmental and evolutionary contexts.
Figures


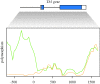

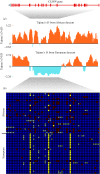
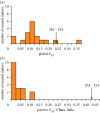

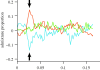
References
-
- Akey J. M., Zhang G., Zhang K., Jin L., Shriver M. D.2002Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12, 1805–1814 (doi:10.1101/gr.631202) - DOI - PMC - PubMed
-
- Akey J. M., Swanson W. J., Madeoy J., Eberle M., Shriver M. D.2006TRPV6 exhibits unusual patterns of polymorphism and divergence in worldwide populations. Hum. Mol. Genet. 15, 2106–2113 (doi:10.1093/hmg/ddl134) - DOI - PubMed
-
- Andres A. M., Soldevila M., Navarro A., Kidd K. K., Oliva B., Bertranpetit J.2004Positive selection in MAOA gene is human exclusive: determination of the putative amino acid change selected in the human lineage. Hum. Genet. 115, 377–386 - PubMed
-
- Ayodo G., Price A. L., Keinan A., Ajwang A., Otieno M. F., Orago A. S., Patterson N., Reich D.2007Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am. J. Hum. Genet. 81, 234–242 (doi:10.1086/519221) - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
