Gene-centered regulatory networks
- PMID: 20008400
- PMCID: PMC3096532
- DOI: 10.1093/bfgp/elp049
Gene-centered regulatory networks
Abstract
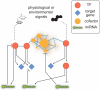
Differential gene expression plays a critical role in the development and physiology of multicellular organisms. At a 'systems level' (e.g. at the level of a tissue, organ or whole organism), this process can be studied using gene regulatory network (GRN) models that capture physical and regulatory interactions between genes and their regulators. In the past years, significant progress has been made toward the mapping of GRNs using a variety of experimental and computational approaches. Here, we will discuss gene-centered approaches that we employed to characterize GRNs and describe insights that we have obtained into the global design principles of gene regulation in complex metazoan systems.
Figures


References
-
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. - PubMed
-
- Gehring WJ, Ikeo K. Pax6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–7. - PubMed
-
- St Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 2009;68:201–19. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

