Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2
- PMID: 20008451
- PMCID: PMC2807926
- DOI: 10.1093/mp/ssp098
Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2
Abstract
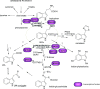
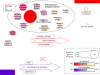
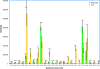
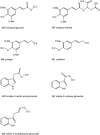
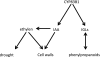
Auxin homeostasis is pivotal for normal plant growth and development. The superroot2 (sur2) mutant was initially isolated in a forward genetic screen for auxin overproducers, and SUR2 was suggested to control auxin conjugation and thereby regulate auxin homeostasis. However, the phenotype was not uniform and could not be described as a pure high auxin phenotype, indicating that knockout of CYP83B1 has multiple effects. Subsequently, SUR2 was identified as CYP83B1, a cytochrome P450 positioned at the metabolic branch point between auxin and indole glucosinolate metabolism. To investigate concomitant global alterations triggered by knockout of CYP83B1 and the countermeasures chosen by the mutant to cope with hormonal and metabolic imbalances, 10-day-old mutant seedlings were characterized with respect to their transcriptome and metabolome profiles. Here, we report a global analysis of the sur2 mutant by the use of a combined transcriptomic and metabolomic approach revealing pronounced effects on several metabolic grids including the intersection between secondary metabolism, cell wall turnover, hormone metabolism, and stress responses. Metabolic and transcriptional cross-talks in sur2 were found to be regulated by complex interactions between both positively and negatively acting transcription factors. The complex phenotype of sur2 may thus not only be assigned to elevated levels of auxin, but also to ethylene and abscisic acid responses as well as drought responses in the absence of a water deficiency. The delicate balance between these signals explains why minute changes in growth conditions may result in the non-uniform phenotype. The large phenotypic variation observed between and within the different surveys may be reconciled by the complex and intricate hormonal balances in sur2 seedlings decoded in this study.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

