MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity
- PMID: 20008554
- PMCID: PMC2815576
- DOI: 10.1128/MCB.01129-09
MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity
Abstract
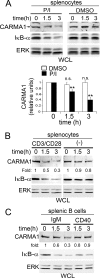
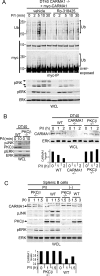
The adaptor protein CARMA1 is required for antigen receptor-triggered activation of IKK and JNK in lymphocytes. Once activated, the events that subsequently turn off the CARMA1 signalosome are unknown. In this study, we found that antigen receptor-activated CARMA1 underwent lysine 48 (K48) polyubiquitination and proteasome-dependent degradation. The MAGUK region of CARMA1 was an essential player in this event; the SH3 and GUK domains contained the main ubiquitin acceptor sites, and deletion of a Hook domain (an important structure for maintaining inactive MAGUK proteins) between SH3 and GUK was sufficient to induce constitutive ubiquitination of CARMA1. A similar deletion promoted the ubiquitination of PSD-95 and Dlgh1, suggesting that a conserved mechanism may control the turnover of other MAGUK family protein complexes. Functionally, we demonstrated that elimination of MAGUK ubiquitination sites in CARMA1 resulted in elevated basal and inducible NF-kappaB and JNK activation as a result of defective K48 ubiquitination and increased persistence of this ubiquitination-deficient CARMA1 protein in activated lymphocytes. The coordination of degradation with the full activation of the CARMA1 molecule likely provides an intrinsic feedback control mechanism to balance lymphocyte activation upon antigenic stimulation.
Figures






References
-
- Bertin, J., L. Wang, Y. Guo, M. D. Jacobson, J. L. Poyet, S. M. Srinivasula, S. Merriam, P. S. DiStefano, and E. S. Alnemri. 2001. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 276:11877-11882. - PubMed
-
- Egawa, T., B. Albrecht, B. Favier, M. J. Sunshine, K. Mirchandani, W. O'Brien, M. Thome, and D. R. Littman. 2003. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr. Biol. 13:1252-1258. - PubMed
-
- Fields, E. R., B. J. Seufzer, E. M. Oltz, and S. Miyamoto. 2000. A switch in distinct I kappa B alpha degradation mechanisms mediates constitutive NF-kappa B activation in mature B cells. J. Immunol. 164:4762-4767. - PubMed
-
- Gaide, O., B. Favier, D. F. Legler, D. Bonnet, B. Brissoni, S. Valitutti, C. Bron, J. Tschopp, and M. Thome. 2002. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat. Immunol. 3:836-843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
