New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans
- PMID: 20008572
- PMCID: PMC2845351
- DOI: 10.1534/genetics.109.110270
New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans
Abstract
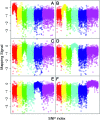
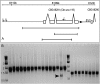
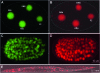
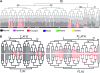
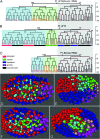
Comparative studies of Caenorhabditis briggsae and C. elegans have provided insights into gene function and developmental control in both organisms. C. elegans is a well developed model organism with a variety of molecular and genetic tools to study gene functions. In contrast, there are only very limited tools available for its closest relative, C. briggsae. To take advantage of the full potential of this comparative approach, we have developed several genetic and molecular tools to facilitate functional analysis in C. briggsae. First, we designed and implemented an SNP-based oligonucleotide microarray for rapid mapping of genetic mutants in C. briggsae. Second, we generated a mutagenized frozen library to permit the isolation of targeted deletions and used the library to recover a deletion mutant of cbr-unc-119 for use as a transgenic marker. Third, we used the cbr-unc-119 mutant in ballistic transformation and generated fluorescently labeled strains that allow automated lineaging and cellular resolution expression analysis. Finally, we demonstrated the potential of automated lineaging by profiling expression of egl-5, hlh-1, and pha-4 at cellular resolution and by detailed phenotyping of the perturbations on the Wnt signaling pathway. These additions to the experimental toolkit for C. briggsae should greatly increase its utility in comparative studies with C. elegans. With the emerging sequence of nematode species more closely related to C. briggsae, these tools may open novel avenues of experimentation in C. briggsae itself.
Figures

References
-
- Ahringer, J., ed. Reverse Genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook. 1.47.1, http://www.wormbook.org.
-
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437 1149–1152. - PubMed
-
- Chen, L., M. Krause, M. Sepanski and A. Fire, 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120 1631–1641. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

