Newly identified antibacterial compounds are topoisomerase poisons in African trypanosomes
- PMID: 20008775
- PMCID: PMC2812133
- DOI: 10.1128/AAC.01025-09
Newly identified antibacterial compounds are topoisomerase poisons in African trypanosomes
Abstract
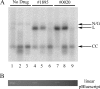
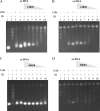
Human African trypanosomiasis, caused by the Trypanosoma brucei protozoan parasite, is fatal when left untreated. Current therapies are antiquated, and there is a need for new pharmacologic agents against T. brucei targets that have no human ortholog. Trypanosomes have a single mitochondrion with a unique mitochondrial DNA, known as kinetoplast DNA (kDNA), a topologically complex network that contains thousands of interlocking circular DNAs, termed minicircles (approximately 1 kb) and maxicircles (approximately 23 kb). Replication of kDNA depends on topoisomerases, enzymes that catalyze reactions that change DNA topology. T. brucei has an unusual type IA topoisomerase that is dedicated to kDNA metabolism. This enzyme has no ortholog in humans, and RNA interference (RNAi) studies have shown that it is essential for parasite survival, making it an ideal drug target. In a large chemical library screen, two compounds were recently identified as poisons of bacterial topoisomerase IA. We found that these compounds are trypanocidal in the low micromolar range and that they promote the formation of linearized minicircles covalently bound to protein on the 5' end, consistent with the poisoning of mitochondrial topoisomerase IA. Surprisingly, however, band depletion studies showed that it is topoisomerase IImt, and not topoisomerase IAmt, that is trapped. Both compounds are planar aromatic polycyclic structures that intercalate into and unwind DNA. These findings reinforce the utility of topoisomerase IImt as a target for development of new drugs for African sleeping sickness.
Figures




Similar articles
-
Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.Nucleic Acids Res. 2017 Aug 21;45(14):8378-8391. doi: 10.1093/nar/gkx521. Nucleic Acids Res. 2017. PMID: 28637278 Free PMC article.
-
A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes.Mol Microbiol. 2008 Feb;67(4):820-9. doi: 10.1111/j.1365-2958.2007.06087.x. Epub 2007 Dec 19. Mol Microbiol. 2008. PMID: 18179422
-
Distinct genes encode type II Topoisomerases for the nucleus and mitochondrion in the protozoan parasite Trypanosoma brucei.J Biol Chem. 2006 Feb 10;281(6):3048-56. doi: 10.1074/jbc.M505977200. Epub 2005 Nov 28. J Biol Chem. 2006. PMID: 16316982
-
Sleeping sickness pathogen (Trypanosoma brucei) and natural products: therapeutic targets and screening systems.Planta Med. 2011 Apr;77(6):586-97. doi: 10.1055/s-0030-1250411. Epub 2010 Oct 13. Planta Med. 2011. PMID: 20945274 Review.
-
The Inhibition of Cysteine Proteases Rhodesain and TbCatB: A Valuable Approach to Treat Human African Trypanosomiasis.Mini Rev Med Chem. 2016;16(17):1374-1391. doi: 10.2174/1389557515666160509125243. Mini Rev Med Chem. 2016. PMID: 27156518 Review.
Cited by
-
Targeting bacterial topoisomerase I to meet the challenge of finding new antibiotics.Future Med Chem. 2015;7(4):459-71. doi: 10.4155/fmc.14.157. Future Med Chem. 2015. PMID: 25875873 Free PMC article. Review.
-
Identification of small molecules with type I interferon inducing properties by high-throughput screening.PLoS One. 2012;7(11):e49049. doi: 10.1371/journal.pone.0049049. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23145065 Free PMC article.
-
Activity of Aromathecins against African Trypanosomes.Antimicrob Agents Chemother. 2018 Oct 24;62(11):e00786-18. doi: 10.1128/AAC.00786-18. Print 2018 Nov. Antimicrob Agents Chemother. 2018. PMID: 30104277 Free PMC article.
-
Old drug repurposing for neglected disease: Pyronaridine as a promising candidate for the treatment of Echinococcus granulosus infections.EBioMedicine. 2020 Apr;54:102711. doi: 10.1016/j.ebiom.2020.102711. Epub 2020 Apr 9. EBioMedicine. 2020. PMID: 32279056 Free PMC article.
-
In vitro trypanocidal activities and structure-activity relationships of ciprofloxacin analogs.Mol Divers. 2024 Aug;28(4):2667-2680. doi: 10.1007/s11030-023-10704-9. Epub 2023 Jul 23. Mol Divers. 2024. PMID: 37481633
References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with Image J. Biophotonics Int. 11:36-42.
-
- Aguero, F., B. Al-Lazikani, M. Aslett, M. Berriman, F. S. Buckner, R. K. Campbell, S. Carmona, I. M. Carruthers, A. W. Chan, F. Chen, G. J. Crowther, M. A. Doyle, C. Hertz-Fowler, A. L. Hopkins, G. McAllister, S. Nwaka, J. P. Overington, A. Pain, G. V. Paolini, U. Pieper, S. A. Ralph, A. Riechers, D. S. Roos, A. Sali, D. Shanmugam, T. Suzuki, W. C. Van Voorhis, and C. L. Verlinde. 2008. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 7:900-907. - PMC - PubMed
-
- Barrett, M. P., R. J. Burchmore, A. Stich, J. O. Lazzari, A. C. Frasch, J. J. Cazzulo, and S. Krishna. 2003. The trypanosomiases. Lancet 362:1469-1480. - PubMed
-
- Bodley, A. L., M. W. McGarry, and T. A. Shapiro. 1995. Drug cytotoxicity assay for African trypanosomes and Leishmania species. J. Infect. Dis. 172:1157-1159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

