miR-19 is a key oncogenic component of mir-17-92
- PMID: 20008935
- PMCID: PMC2800084
- DOI: 10.1101/gad.1861409
miR-19 is a key oncogenic component of mir-17-92
Abstract
Recent studies have revealed the importance of multiple microRNAs (miRNAs) in promoting tumorigenesis, among which mir-17-92/Oncomir-1 exhibits potent oncogenic activity. Genomic amplification and elevated expression of mir-17-92 occur in several human B-cell lymphomas, and enforced mir-17-92 expression in mice cooperates with c-myc to promote the formation of B-cell lymphomas. Unlike classic protein-coding oncogenes, mir-17-92 has an unconventional gene structure, where one primary transcript yields six individual miRNAs. Here, we functionally dissected the individual components of mir-17-92 by assaying their tumorigenic potential in vivo. Using the Emu-myc model of mouse B-cell lymphoma, we identified miR-19 as the key oncogenic component of mir-17-92, both necessary and sufficient for promoting c-myc-induced lymphomagenesis by repressing apoptosis. The oncogenic activity of miR-19 is at least in part due to its repression of the tumor suppressor Pten. Consistently, miR-19 activates the Akt-mTOR (mammalian target of rapamycin) pathway, thereby functionally antagonizing Pten to promote cell survival. Our findings reveal the essential role of miR-19 in mediating the oncogenic activity of mir-17-92, and implicate the functional diversity of mir-17-92 components as the molecular basis for its pleiotropic effects during tumorigenesis.
Figures

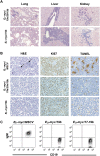
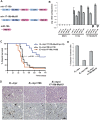
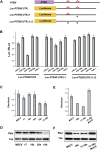


Comment in
-
Tumorigenicity of the miR-17-92 cluster distilled.Genes Dev. 2010 Jan 1;24(1):1-4. doi: 10.1101/gad.1887110. Genes Dev. 2010. PMID: 20047995 Free PMC article.
Similar articles
-
A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis.Elife. 2013 Oct 15;2:e00822. doi: 10.7554/eLife.00822. Elife. 2013. PMID: 24137534 Free PMC article.
-
The PTEN-AKT-mTOR/RICTOR Pathway in Nasal Natural Killer Cell Lymphoma Is Activated by miR-494-3p via PTEN But Inhibited by miR-142-3p via RICTOR.Am J Pathol. 2015 May;185(5):1487-99. doi: 10.1016/j.ajpath.2015.01.025. Am J Pathol. 2015. PMID: 25907832
-
miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer.Cancer Res. 2013 Sep 1;73(17):5402-15. doi: 10.1158/0008-5472.CAN-13-0297. Epub 2013 Jul 15. Cancer Res. 2013. PMID: 23856247
-
mir-17-92, a cluster of miRNAs in the midst of the cancer network.Int J Biochem Cell Biol. 2010 Aug;42(8):1348-54. doi: 10.1016/j.biocel.2010.03.004. Epub 2010 Mar 19. Int J Biochem Cell Biol. 2010. PMID: 20227518 Free PMC article. Review.
-
mir-17-92: a polycistronic oncomir with pleiotropic functions.Immunol Rev. 2013 May;253(1):158-66. doi: 10.1111/imr.12054. Immunol Rev. 2013. PMID: 23550645 Free PMC article. Review.
Cited by
-
Reprogramming of the microRNA transcriptome mediates resistance to rapamycin.J Biol Chem. 2013 Mar 1;288(9):6034-44. doi: 10.1074/jbc.M112.416446. Epub 2013 Jan 8. J Biol Chem. 2013. PMID: 23300087 Free PMC article.
-
A comprehensive in vivo screen for anti-apoptotic miRNAs indicates broad capacities for oncogenic synergy.Dev Biol. 2021 Jul;475:10-20. doi: 10.1016/j.ydbio.2021.02.010. Epub 2021 Mar 1. Dev Biol. 2021. PMID: 33662357 Free PMC article.
-
NOTCH and EZH2 collaborate to repress PTEN expression in breast cancer.Commun Biol. 2021 Mar 9;4(1):312. doi: 10.1038/s42003-021-01825-8. Commun Biol. 2021. PMID: 33750924 Free PMC article.
-
Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis.Cancer Cell. 2012 Aug 14;22(2):167-79. doi: 10.1016/j.ccr.2012.06.012. Cancer Cell. 2012. PMID: 22897848 Free PMC article.
-
Neuroprotection induced by post-conditioning following ischemia/reperfusion in mice is associated with altered microRNA expression.Mol Med Rep. 2016 Sep;14(3):2582-8. doi: 10.3892/mmr.2016.5576. Epub 2016 Jul 29. Mol Med Rep. 2016. PMID: 27485299 Free PMC article.
References
-
- Abubaker J, Bavi PP, Al-Harbi S, Siraj AK, Al-Dayel F, Uddin S, Al-Kuraya K. PIK3CA mutations are mutually exclusive with PTEN loss in diffuse large B-cell lymphoma. Leukemia. 2007;21:2368–2370. - PubMed
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
