Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3' regulatory region
- PMID: 20010689
- PMCID: PMC2802177
- DOI: 10.1038/nature08633
Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3' regulatory region
Abstract
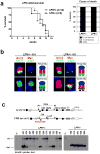
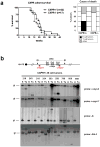
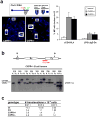
B-cell malignancies, such as human Burkitt's lymphoma, often contain translocations that link c-myc or other proto-oncogenes to the immunoglobulin heavy chain locus (IgH, encoded by Igh). The nature of elements that activate oncogenes within such translocations has been a long-standing question. Translocations within Igh involve DNA double-strand breaks initiated either by the RAG1/2 endonuclease during variable, diversity and joining gene segment (V(D)J) recombination, or by activation-induced cytidine deaminase (AID, also known as AICDA) during class switch recombination (CSR). V(D)J recombination in progenitor B (pro-B) cells assembles Igh variable region exons upstream of mu constant region (Cmu) exons, which are the first of several sets of C(H) exons ('C(H) genes') within a C(H) locus that span several hundred kilobases (kb). In mature B cells, CSR deletes Cmu and replaces it with a downstream C(H) gene. An intronic enhancer (iEmu) between the variable region exons and Cmu promotes V(D)J recombination in developing B cells. Furthermore, the Igh 3' regulatory region (Igh3'RR) lies downstream of the C(H) locus and modulates CSR by long-range transcriptional enhancement of C(H) genes. Transgenic mice bearing iEmu or Igh3'RR sequences fused to c-myc are predisposed to B lymphomas, demonstrating that such elements can confer oncogenic c-myc expression. However, in many B-cell lymphomas, Igh-c-myc translocations delete iEmu and place c-myc up to 200 kb upstream of the Igh3'RR. Here we address the oncogenic role of the Igh3'RR by inactivating it in two distinct mouse models for B-cell lymphoma with Igh-c-myc translocations. We show that the Igh3'RR is dispensable for pro-B-cell lymphomas with V(D)J recombination-initiated translocations, but is required for peripheral B-cell lymphomas with CSR-associated translocations. As the Igh3'RR is not required for CSR-associated Igh breaks or Igh-c-myc translocations in peripheral B-cell lymphoma progenitors, we conclude that this regulatory region confers oncogenic activity by long-range and developmental stage-specific activation of translocated c-myc genes.
Figures

Similar articles
-
Mechanisms promoting translocations in editing and switching peripheral B cells.Nature. 2009 Jul 9;460(7252):231-6. doi: 10.1038/nature08159. Nature. 2009. PMID: 19587764 Free PMC article.
-
Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes.Proc Natl Acad Sci U S A. 2014 Jul 15;111(28):10269-74. doi: 10.1073/pnas.1410112111. Epub 2014 Jun 30. Proc Natl Acad Sci U S A. 2014. PMID: 24982162 Free PMC article.
-
IgH class switching and translocations use a robust non-classical end-joining pathway.Nature. 2007 Sep 27;449(7161):478-82. doi: 10.1038/nature06020. Epub 2007 Aug 22. Nature. 2007. PMID: 17713479
-
Myc translocations in B cell and plasma cell neoplasms.DNA Repair (Amst). 2006 Sep 8;5(9-10):1213-24. doi: 10.1016/j.dnarep.2006.05.017. Epub 2006 Jul 11. DNA Repair (Amst). 2006. PMID: 16815105 Review.
-
Mouse Models of c-myc Deregulation Driven by IgH Locus Enhancers as Models of B-Cell Lymphomagenesis.Front Immunol. 2020 Jul 23;11:1564. doi: 10.3389/fimmu.2020.01564. eCollection 2020. Front Immunol. 2020. PMID: 32793219 Free PMC article. Review.
Cited by
-
Genome instability and lymphoma.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021 May 28;46(5):552-557. doi: 10.11817/j.issn.1672-7347.2021.190427. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34148893 Free PMC article. Chinese, English.
-
Origin of chromosomal translocations in lymphoid cancer.Cell. 2010 Apr 2;141(1):27-38. doi: 10.1016/j.cell.2010.03.016. Cell. 2010. PMID: 20371343 Free PMC article. Review.
-
Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by Topo-isomerase I inhibitors.Elife. 2016 Jun 15;5:e14709. doi: 10.7554/eLife.14709. Elife. 2016. PMID: 27304073 Free PMC article.
-
Mechanisms that can promote peripheral B-cell lymphoma in ATM-deficient mice.Cancer Immunol Res. 2014 Sep;2(9):857-66. doi: 10.1158/2326-6066.CIR-14-0090. Epub 2014 Jun 9. Cancer Immunol Res. 2014. PMID: 24913718 Free PMC article.
-
The IgH 3' regulatory region and c-myc-induced B-cell lymphomagenesis.Oncotarget. 2017 Jan 24;8(4):7059-7067. doi: 10.18632/oncotarget.12535. Oncotarget. 2017. PMID: 27729620 Free PMC article. Review.
References
-
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. - PubMed
-
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. - PubMed
-
- Ramiro A, et al. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv Immunol. 2007;94:75–107. - PubMed
-
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

