The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes
- PMID: 20011103
- PMCID: PMC2781106
- DOI: 10.1371/journal.pcbi.1000593
The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes
Abstract
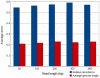
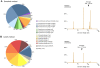
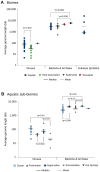
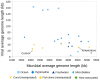
Metagenomic studies characterize both the composition and diversity of uncultured viral and microbial communities. BLAST-based comparisons have typically been used for such analyses; however, sampling biases, high percentages of unknown sequences, and the use of arbitrary thresholds to find significant similarities can decrease the accuracy and validity of estimates. Here, we present Genome relative Abundance and Average Size (GAAS), a complete software package that provides improved estimates of community composition and average genome length for metagenomes in both textual and graphical formats. GAAS implements a novel methodology to control for sampling bias via length normalization, to adjust for multiple BLAST similarities by similarity weighting, and to select significant similarities using relative alignment lengths. In benchmark tests, the GAAS method was robust to both high percentages of unknown sequences and to variations in metagenomic sequence read lengths. Re-analysis of the Sargasso Sea virome using GAAS indicated that standard methodologies for metagenomic analysis may dramatically underestimate the abundance and importance of organisms with small genomes in environmental systems. Using GAAS, we conducted a meta-analysis of microbial and viral average genome lengths in over 150 metagenomes from four biomes to determine whether genome lengths vary consistently between and within biomes, and between microbial and viral communities from the same environment. Significant differences between biomes and within aquatic sub-biomes (oceans, hypersaline systems, freshwater, and microbialites) suggested that average genome length is a fundamental property of environments driven by factors at the sub-biome level. The behavior of paired viral and microbial metagenomes from the same environment indicated that microbial and viral average genome sizes are independent of each other, but indicative of community responses to stressors and environmental conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Quantitative metagenomic analyses based on average genome size normalization.Appl Environ Microbiol. 2011 Apr;77(7):2513-21. doi: 10.1128/AEM.02167-10. Epub 2011 Feb 11. Appl Environ Microbiol. 2011. PMID: 21317268 Free PMC article.
-
Metagenomic signatures of 86 microbial and viral metagenomes.Environ Microbiol. 2009 Jul;11(7):1752-66. doi: 10.1111/j.1462-2920.2009.01901.x. Epub 2009 Mar 18. Environ Microbiol. 2009. PMID: 19302541
-
Assessing the diversity and specificity of two freshwater viral communities through metagenomics.PLoS One. 2012;7(3):e33641. doi: 10.1371/journal.pone.0033641. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432038 Free PMC article.
-
From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems.Cell Mol Life Sci. 2015 Nov;72(22):4287-308. doi: 10.1007/s00018-015-2004-1. Epub 2015 Aug 9. Cell Mol Life Sci. 2015. PMID: 26254872 Free PMC article. Review.
-
Applying Genome-Resolved Metagenomics to Deconvolute the Halophilic Microbiome.Genes (Basel). 2019 Mar 14;10(3):220. doi: 10.3390/genes10030220. Genes (Basel). 2019. PMID: 30875864 Free PMC article. Review.
Cited by
-
Purifying the impure: sequencing metagenomes and metatranscriptomes from complex animal-associated samples.J Vis Exp. 2014 Dec 22;(94):52117. doi: 10.3791/52117. J Vis Exp. 2014. PMID: 25549184 Free PMC article.
-
Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis.ISME J. 2015 Oct;9(10):2162-77. doi: 10.1038/ismej.2015.28. Epub 2015 Jun 30. ISME J. 2015. PMID: 26125684 Free PMC article.
-
Bacterial niche-specific genome expansion is coupled with highly frequent gene disruptions in deep-sea sediments.PLoS One. 2011;6(12):e29149. doi: 10.1371/journal.pone.0029149. Epub 2011 Dec 21. PLoS One. 2011. PMID: 22216192 Free PMC article.
-
MLTreeMap--accurate Maximum Likelihood placement of environmental DNA sequences into taxonomic and functional reference phylogenies.BMC Genomics. 2010 Aug 5;11:461. doi: 10.1186/1471-2164-11-461. BMC Genomics. 2010. PMID: 20687950 Free PMC article.
-
CoreProbe: A Novel Algorithm for Estimating Relative Abundance Based on Metagenomic Reads.Genes (Basel). 2018 Jun 20;9(6):313. doi: 10.3390/genes9060313. Genes (Basel). 2018. PMID: 29925824 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

