Challenges predicting ligand-receptor interactions of promiscuous proteins: the nuclear receptor PXR
- PMID: 20011107
- PMCID: PMC2781111
- DOI: 10.1371/journal.pcbi.1000594
Challenges predicting ligand-receptor interactions of promiscuous proteins: the nuclear receptor PXR
Abstract
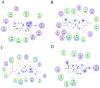
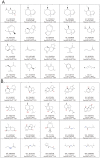
Transcriptional regulation of some genes involved in xenobiotic detoxification and apoptosis is performed via the human pregnane X receptor (PXR) which in turn is activated by structurally diverse agonists including steroid hormones. Activation of PXR has the potential to initiate adverse effects, altering drug pharmacokinetics or perturbing physiological processes. Reliable computational prediction of PXR agonists would be valuable for pharmaceutical and toxicological research. There has been limited success with structure-based modeling approaches to predict human PXR activators. Slightly better success has been achieved with ligand-based modeling methods including quantitative structure-activity relationship (QSAR) analysis, pharmacophore modeling and machine learning. In this study, we present a comprehensive analysis focused on prediction of 115 steroids for ligand binding activity towards human PXR. Six crystal structures were used as templates for docking and ligand-based modeling approaches (two-, three-, four- and five-dimensional analyses). The best success at external prediction was achieved with 5D-QSAR. Bayesian models with FCFP_6 descriptors were validated after leaving a large percentage of the dataset out and using an external test set. Docking of ligands to the PXR structure co-crystallized with hyperforin had the best statistics for this method. Sulfated steroids (which are activators) were consistently predicted as non-activators while, poorly predicted steroids were docked in a reverse mode compared to 5alpha-androstan-3beta-ol. Modeling of human PXR represents a complex challenge by virtue of the large, flexible ligand-binding cavity. This study emphasizes this aspect, illustrating modest success using the largest quantitative data set to date and multiple modeling approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Ligand- and structure-based pregnane X receptor models.Methods Mol Biol. 2012;929:359-75. doi: 10.1007/978-1-62703-050-2_15. Methods Mol Biol. 2012. PMID: 23007437 Review.
-
Machine learning and traditional QSAR modeling methods: a case study of known PXR activators.J Biomol Struct Dyn. 2024 Jan-Feb;42(2):903-917. doi: 10.1080/07391102.2023.2196701. Epub 2023 Apr 14. J Biomol Struct Dyn. 2024. PMID: 37059719
-
Hybrid scoring and classification approaches to predict human pregnane X receptor activators.Pharm Res. 2009 Apr;26(4):1001-11. doi: 10.1007/s11095-008-9809-7. Epub 2008 Dec 30. Pharm Res. 2009. PMID: 19115096 Free PMC article.
-
Development of in silico filters to predict activation of the pregnane X receptor (PXR) by structurally diverse drug-like molecules.Bioorg Med Chem. 2012 Sep 15;20(18):5352-65. doi: 10.1016/j.bmc.2012.04.020. Epub 2012 Apr 19. Bioorg Med Chem. 2012. PMID: 22560839
-
Functional evolution of the pregnane X receptor.Expert Opin Drug Metab Toxicol. 2006 Jun;2(3):381-97. doi: 10.1517/17425255.2.3.381. Expert Opin Drug Metab Toxicol. 2006. PMID: 16863441 Free PMC article. Review.
Cited by
-
Novel yeast-based strategy unveils antagonist binding regions on the nuclear xenobiotic receptor PXR.J Biol Chem. 2013 May 10;288(19):13655-68. doi: 10.1074/jbc.M113.455485. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525103 Free PMC article.
-
In silico investigation of agonist activity of a structurally diverse set of drugs to hPXR using HM-BSM and HM-PNN.J Huazhong Univ Sci Technolog Med Sci. 2016 Jun;36(3):463-468. doi: 10.1007/s11596-016-1609-4. Epub 2016 Jul 5. J Huazhong Univ Sci Technolog Med Sci. 2016. PMID: 27376821
-
Drug Mimicry: Promiscuous Receptors PXR and AhR, and Microbial Metabolite Interactions in the Intestine.Trends Pharmacol Sci. 2020 Dec;41(12):900-908. doi: 10.1016/j.tips.2020.09.013. Epub 2020 Oct 20. Trends Pharmacol Sci. 2020. PMID: 33097284 Free PMC article. Review.
-
Exploring the PXR ligand binding mechanism with advanced Molecular Dynamics methods.Sci Rep. 2018 Nov 1;8(1):16207. doi: 10.1038/s41598-018-34373-z. Sci Rep. 2018. PMID: 30385820 Free PMC article.
-
Back to the future: Advances in development of broad-spectrum capsid-binding inhibitors of enteroviruses.Eur J Med Chem. 2019 Sep 15;178:606-622. doi: 10.1016/j.ejmech.2019.06.008. Epub 2019 Jun 11. Eur J Med Chem. 2019. PMID: 31226653 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

