Annotation error in public databases: misannotation of molecular function in enzyme superfamilies
- PMID: 20011109
- PMCID: PMC2781113
- DOI: 10.1371/journal.pcbi.1000605
Annotation error in public databases: misannotation of molecular function in enzyme superfamilies
Abstract
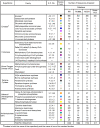
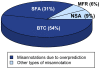
Due to the rapid release of new data from genome sequencing projects, the majority of protein sequences in public databases have not been experimentally characterized; rather, sequences are annotated using computational analysis. The level of misannotation and the types of misannotation in large public databases are currently unknown and have not been analyzed in depth. We have investigated the misannotation levels for molecular function in four public protein sequence databases (UniProtKB/Swiss-Prot, GenBank NR, UniProtKB/TrEMBL, and KEGG) for a model set of 37 enzyme families for which extensive experimental information is available. The manually curated database Swiss-Prot shows the lowest annotation error levels (close to 0% for most families); the two other protein sequence databases (GenBank NR and TrEMBL) and the protein sequences in the KEGG pathways database exhibit similar and surprisingly high levels of misannotation that average 5%-63% across the six superfamilies studied. For 10 of the 37 families examined, the level of misannotation in one or more of these databases is >80%. Examination of the NR database over time shows that misannotation has increased from 1993 to 2005. The types of misannotation that were found fall into several categories, most associated with "overprediction" of molecular function. These results suggest that misannotation in enzyme superfamilies containing multiple families that catalyze different reactions is a larger problem than has been recognized. Strategies are suggested for addressing some of the systematic problems contributing to these high levels of misannotation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Bork P, Bairoch A. Go hunting in sequence databases but watch out for the traps. Trends Genet. 1996;12:425–427. - PubMed
-
- Karp PD. What we do not know about sequence analysis and sequence databases. Bioinformatics. 1998;14:753–754. - PubMed
-
- Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. - PubMed
-
- Devos D, Valencia A. Intrinsic errors in genome annotation. Trends Genet. 2001;17:429–431. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

