Early evolution of conserved regulatory sequences associated with development in vertebrates
- PMID: 20011110
- PMCID: PMC2781166
- DOI: 10.1371/journal.pgen.1000762
Early evolution of conserved regulatory sequences associated with development in vertebrates
Abstract
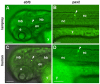
Comparisons between diverse vertebrate genomes have uncovered thousands of highly conserved non-coding sequences, an increasing number of which have been shown to function as enhancers during early development. Despite their extreme conservation over 500 million years from humans to cartilaginous fish, these elements appear to be largely absent in invertebrates, and, to date, there has been little understanding of their mode of action or the evolutionary processes that have modelled them. We have now exploited emerging genomic sequence data for the sea lamprey, Petromyzon marinus, to explore the depth of conservation of this type of element in the earliest diverging extant vertebrate lineage, the jawless fish (agnathans). We searched for conserved non-coding elements (CNEs) at 13 human gene loci and identified lamprey elements associated with all but two of these gene regions. Although markedly shorter and less well conserved than within jawed vertebrates, identified lamprey CNEs are able to drive specific patterns of expression in zebrafish embryos, which are almost identical to those driven by the equivalent human elements. These CNEs are therefore a unique and defining characteristic of all vertebrates. Furthermore, alignment of lamprey and other vertebrate CNEs should permit the identification of persistent sequence signatures that are responsible for common patterns of expression and contribute to the elucidation of the regulatory language in CNEs. Identifying the core regulatory code for development, common to all vertebrates, provides a foundation upon which regulatory networks can be constructed and might also illuminate how large conserved regulatory sequence blocks evolve and become fixed in genomic DNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A reporter assay in lamprey embryos reveals both functional conservation and elaboration of vertebrate enhancers.PLoS One. 2014 Jan 9;9(1):e85492. doi: 10.1371/journal.pone.0085492. eCollection 2014. PLoS One. 2014. PMID: 24416417 Free PMC article.
-
A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates.Nature. 2014 Oct 23;514(7523):490-3. doi: 10.1038/nature13723. Epub 2014 Sep 14. Nature. 2014. PMID: 25219855 Free PMC article.
-
Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum).Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):16044-9. doi: 10.1073/pnas.1315760110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043829 Free PMC article.
-
Functional genetic analysis in a jawless vertebrate, the sea lamprey: insights into the developmental evolution of early vertebrates.J Exp Biol. 2020 Feb 7;223(Pt Suppl 1):jeb206433. doi: 10.1242/jeb.206433. J Exp Biol. 2020. PMID: 32034037 Review.
-
Landmark discoveries in elucidating the origins of the hypothalamic-pituitary system from the perspective of a basal vertebrate, sea lamprey.Gen Comp Endocrinol. 2018 Aug 1;264:3-15. doi: 10.1016/j.ygcen.2017.10.016. Epub 2017 Oct 27. Gen Comp Endocrinol. 2018. PMID: 29111305 Review.
Cited by
-
An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates.Dev Biol. 2019 Sep 1;453(1):19-33. doi: 10.1016/j.ydbio.2019.05.001. Epub 2019 May 6. Dev Biol. 2019. PMID: 31071313 Free PMC article.
-
The mystery of extreme non-coding conservation.Philos Trans R Soc Lond B Biol Sci. 2013 Nov 11;368(1632):20130021. doi: 10.1098/rstb.2013.0021. Print 2013 Dec 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24218634 Free PMC article. Review.
-
Heterogeneous conservation of Dlx paralog co-expression in jawed vertebrates.PLoS One. 2013 Jun 28;8(6):e68182. doi: 10.1371/journal.pone.0068182. Print 2013. PLoS One. 2013. PMID: 23840829 Free PMC article.
-
Cellular and Molecular Features of Developmentally Programmed Genome Rearrangement in a Vertebrate (Sea Lamprey: Petromyzon marinus).PLoS Genet. 2016 Jun 24;12(6):e1006103. doi: 10.1371/journal.pgen.1006103. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27341395 Free PMC article.
-
Vertebrate paralogous conserved noncoding sequences may be related to gene expressions in brain.Genome Biol Evol. 2013;5(1):140-50. doi: 10.1093/gbe/evs128. Genome Biol Evol. 2013. PMID: 23267051 Free PMC article.
References
-
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. - DOI - PMC - PubMed
-
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, et al. Ancient noncoding elements conserved in the human genome. Science. 2006;314:1892. - PubMed
-
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. - PubMed
-
- Pennacchio L, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

