Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression
- PMID: 20011120
- PMCID: PMC2782133
- DOI: 10.1371/journal.pgen.1000769
Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression
Abstract
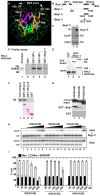
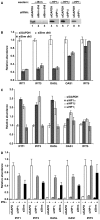
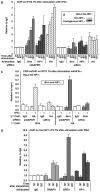
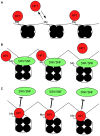
The heterochromatin-enriched HP1 proteins play a critical role in regulation of transcription. These proteins contain two related domains known as the chromo- and the chromoshadow-domain. The chromo-domain binds histone H3 tails methylated on lysine 9. However, in vivo and in vitro experiments have shown that the affinity of HP1 proteins to native methylated chromatin is relatively poor and that the opening of chromatin occurring during DNA replication facilitates their binding to nucleosomes. These observations prompted us to investigate whether HP1 proteins have additional histone binding activities, envisioning also affinity for regions potentially occluded by the nucleosome structure. We find that the chromoshadow-domain interacts with histone H3 in a region located partially inside the nucleosomal barrel at the entry/exit point of the nucleosome. Interestingly, this region is also contacted by the catalytic subunits of the human SWI/SNF complex. In vitro, efficient SWI/SNF remodeling requires this contact and is inhibited in the presence of HP1 proteins. The antagonism between SWI/SNF and HP1 proteins is also observed in vivo on a series of interferon-regulated genes. Finally, we show that SWI/SNF activity favors loading of HP1 proteins to chromatin both in vivo and in vitro. Altogether, our data suggest that HP1 chromoshadow-domains can benefit from the opening of nucleosomal structures to bind chromatin and that HP1 proteins use this property to detect and arrest unwanted chromatin remodeling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article.
-
Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain.Nature. 2001 Mar 1;410(6824):120-4. doi: 10.1038/35065138. Nature. 2001. PMID: 11242054
-
Functional selectivity of recombinant mammalian SWI/SNF subunits.Genes Dev. 2000 Oct 1;14(19):2441-51. doi: 10.1101/gad.828000. Genes Dev. 2000. PMID: 11018012 Free PMC article.
-
How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review.
-
Chromatin remodeling during glucocorticoid receptor regulated transactivation.Biochim Biophys Acta. 2012 Jul;1819(7):716-26. doi: 10.1016/j.bbagrm.2012.02.019. Epub 2012 Mar 6. Biochim Biophys Acta. 2012. PMID: 22425674 Free PMC article. Review.
Cited by
-
Heterochromatin protein 1 alpha (HP1α) undergoes a monomer to dimer transition that opens and compacts live cell genome architecture.Nucleic Acids Res. 2024 Oct 14;52(18):10918-10933. doi: 10.1093/nar/gkae720. Nucleic Acids Res. 2024. PMID: 39193905 Free PMC article.
-
Shigella flexneri targets the HP1γ subcode through the phosphothreonine lyase OspF.EMBO J. 2014 Nov 18;33(22):2606-22. doi: 10.15252/embj.201489244. Epub 2014 Sep 12. EMBO J. 2014. PMID: 25216677 Free PMC article.
-
New twists of a TAIL: novel insights into the histone binding properties of a highly conserved PHD finger cluster within the MLR family of H3K4 mono-methyltransferases.Nucleic Acids Res. 2023 Oct 13;51(18):9672-9689. doi: 10.1093/nar/gkad698. Nucleic Acids Res. 2023. PMID: 37638761 Free PMC article.
-
Phosphorylated HP1α-Nucleosome Interactions in Phase Separated Environments.J Am Chem Soc. 2023 Nov 8;145(44):23994-24004. doi: 10.1021/jacs.3c06481. Epub 2023 Oct 23. J Am Chem Soc. 2023. PMID: 37870432 Free PMC article.
-
A new, highly conserved domain in Swi2/Snf2 is required for SWI/SNF remodeling.Nucleic Acids Res. 2011 Nov;39(21):9155-66. doi: 10.1093/nar/gkr622. Epub 2011 Aug 10. Nucleic Acids Res. 2011. PMID: 21835776 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

