Cytoplasmic compartmentalization of the fetal piRNA pathway in mice
- PMID: 20011505
- PMCID: PMC2785470
- DOI: 10.1371/journal.pgen.1000764
Cytoplasmic compartmentalization of the fetal piRNA pathway in mice
Abstract
Derepression of transposable elements (TEs) in the course of epigenetic reprogramming of the mouse embryonic germline necessitates the existence of a robust defense that is comprised of PIWI/piRNA pathway and de novo DNA methylation machinery. To gain further insight into biogenesis and function of piRNAs, we studied the intracellular localization of piRNA pathway components and used the combination of genetic, molecular, and cell biological approaches to examine the performance of the piRNA pathway in germ cells of mice lacking Maelstrom (MAEL), an evolutionarily conserved protein implicated in transposon silencing in fruit flies and mice. Here we show that principal components of the fetal piRNA pathway, MILI and MIWI2 proteins, localize to two distinct types of germinal cytoplasmic granules and exhibit differential association with components of the mRNA degradation/translational repression machinery. The first type of granules, pi-bodies, contains the MILI-TDRD1 module of the piRNA pathway and is likely equivalent to the enigmatic "cementing material" first described in electron micrographs of rat gonocytes over 35 years ago. The second type of granules, piP-bodies, harbors the MIWI2-TDRD9-MAEL module of the piRNA pathway and signature components of P-bodies, GW182, DCP1a, DDX6/p54, and XRN1 proteins. piP-bodies are found predominantly in the proximity of pi-bodies and the two frequently share mouse VASA homolog (MVH) protein, an RNA helicase. In Mael-mutant gonocytes, MIWI2, TDRD9, and MVH are lost from piP-bodies, whereas no effects on pi-body composition are observed. Further analysis revealed that MAEL appears to specifically facilitate MIWI2-dependent aspects of the piRNA pathway including biogenesis of secondary piRNAs, de novo DNA methylation, and efficient downregulation of TEs. Cumulatively, our data reveal elaborate cytoplasmic compartmentalization of the fetal piRNA pathway that relies on MAEL function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
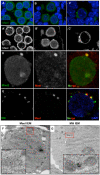
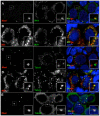
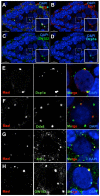

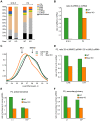

Comment in
-
Retrotransposon silencing by piRNAs: ping-pong players mark their sub-cellular boundaries.PLoS Genet. 2009 Dec;5(12):e1000770. doi: 10.1371/journal.pgen.1000770. Epub 2009 Dec 11. PLoS Genet. 2009. PMID: 20011121 Free PMC article. No abstract available.
References
-
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

