rRNA maturation in yeast cells depleted of large ribosomal subunit proteins
- PMID: 20011513
- PMCID: PMC2788216
- DOI: 10.1371/journal.pone.0008249
rRNA maturation in yeast cells depleted of large ribosomal subunit proteins
Abstract
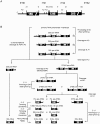
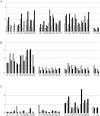
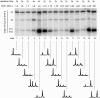
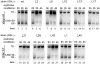
The structural constituents of the large eukaryotic ribosomal subunit are 3 ribosomal RNAs, namely the 25S, 5.8S and 5S rRNA and about 46 ribosomal proteins (r-proteins). They assemble and mature in a highly dynamic process that involves more than 150 proteins and 70 small RNAs. Ribosome biogenesis starts in the nucleolus, continues in the nucleoplasm and is completed after nucleo-cytoplasmic translocation of the subunits in the cytoplasm. In this work we created 26 yeast strains, each of which conditionally expresses one of the large ribosomal subunit (LSU) proteins. In vivo depletion of the analysed LSU r-proteins was lethal and led to destabilisation and degradation of the LSU and/or its precursors. Detailed steady state and metabolic pulse labelling analyses of rRNA precursors in these mutant strains showed that LSU r-proteins can be grouped according to their requirement for efficient progression of different steps of large ribosomal subunit maturation. Comparative analyses of the observed phenotypes and the nature of r-protein-rRNA interactions as predicted by current atomic LSU structure models led us to discuss working hypotheses on i) how individual r-proteins control the productive processing of the major 5' end of 5.8S rRNA precursors by exonucleases Rat1p and Xrn1p, and ii) the nature of structural characteristics of nascent LSUs that are required for cytoplasmic accumulation of nascent subunits but are nonessential for most of the nuclear LSU pre-rRNA processing events.
Conflict of interest statement
Figures

Similar articles
-
Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae.RNA Biol. 2015;12(8):838-46. doi: 10.1080/15476286.2015.1058477. RNA Biol. 2015. PMID: 26151772 Free PMC article.
-
ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs.Nucleic Acids Res. 2001 Sep 1;29(17):3621-30. doi: 10.1093/nar/29.17.3621. Nucleic Acids Res. 2001. PMID: 11522832 Free PMC article.
-
The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae.Mol Cell Biol. 2010 Feb;30(4):976-84. doi: 10.1128/MCB.01359-09. Epub 2009 Dec 14. Mol Cell Biol. 2010. PMID: 20008552 Free PMC article.
-
RNA folding and functions of RNA helicases in ribosome biogenesis.RNA Biol. 2022 Jan;19(1):781-810. doi: 10.1080/15476286.2022.2079890. RNA Biol. 2022. PMID: 35678541 Free PMC article. Review.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
-
Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae.Nucleic Acids Res. 2013 Jan;41(2):1191-210. doi: 10.1093/nar/gks1056. Epub 2012 Dec 2. Nucleic Acids Res. 2013. PMID: 23209026 Free PMC article.
-
Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae.PLoS One. 2013 Jul 10;8(7):e68412. doi: 10.1371/journal.pone.0068412. Print 2013. PLoS One. 2013. PMID: 23874617 Free PMC article.
-
Wide mutational analysis to ascertain the functional roles of eL33 in ribosome biogenesis and translation initiation.Curr Genet. 2022 Dec;68(5-6):619-644. doi: 10.1007/s00294-022-01251-1. Epub 2022 Aug 22. Curr Genet. 2022. PMID: 35994100 Free PMC article.
-
Tsr4 Is a Cytoplasmic Chaperone for the Ribosomal Protein Rps2 in Saccharomyces cerevisiae.Mol Cell Biol. 2019 Aug 12;39(17):e00094-19. doi: 10.1128/MCB.00094-19. Print 2019 Sep 1. Mol Cell Biol. 2019. PMID: 31182640 Free PMC article.
-
Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA.Nucleic Acids Res. 2013 Jan;41(2):1151-63. doi: 10.1093/nar/gks1102. Epub 2012 Nov 23. Nucleic Acids Res. 2013. PMID: 23180764 Free PMC article.
References
-
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. - DOI - PMC - PubMed
-
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. - DOI - PMC - PubMed
-
- Liang X, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. 2009 RNA Available: http://www.ncbi.nlm.nih.gov/pubmed/19628622. Accessed 14 Aug 2009. - PMC - PubMed
-
- Liang X, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. - DOI - PubMed
-
- Ferreira-Cerca S, Pöll G, Gleizes P, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

