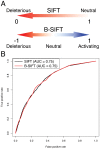
Bi-directional SIFT predicts a subset of activating mutations
- PMID: 20011534
- PMCID: PMC2788704
- DOI: 10.1371/journal.pone.0008311
Bi-directional SIFT predicts a subset of activating mutations
Abstract
Advancements in sequencing technologies have empowered recent efforts to identify polymorphisms and mutations on a global scale. The large number of variations and mutations found in these projects requires high-throughput tools to identify those that are most likely to have an impact on function. Numerous computational tools exist for predicting which mutations are likely to be functional, but none that specifically attempt to identify mutations that result in hyperactivation or gain-of-function. Here we present a modified version of the SIFT (Sorting Intolerant from Tolerant) algorithm that utilizes protein sequence alignments with homologous sequences to identify functional mutations based on evolutionary fitness. We show that this bi-directional SIFT (B-SIFT) is capable of identifying experimentally verified activating mutants from multiple datasets. B-SIFT analysis of large-scale cancer genotyping data identified potential activating mutations, some of which we have provided detailed structural evidence to support. B-SIFT could prove to be a valuable tool for efforts in protein engineering as well as in identification of functional mutations in cancer.
Conflict of interest statement
Figures


Similar articles
-
Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations.Genet Test Mol Biomarkers. 2010 Aug;14(4):533-7. doi: 10.1089/gtmb.2010.0036. Genet Test Mol Biomarkers. 2010. PMID: 20642364
-
Distinguishing cancer-associated missense mutations from common polymorphisms.Cancer Res. 2007 Jan 15;67(2):465-73. doi: 10.1158/0008-5472.CAN-06-1736. Cancer Res. 2007. PMID: 17234753
-
In silico searching for disease-associated functional DNA variants.Methods Mol Biol. 2011;760:239-50. doi: 10.1007/978-1-61779-176-5_15. Methods Mol Biol. 2011. PMID: 21780001
-
Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes.Neuroscience. 2007 Apr 14;145(4):1273-9. doi: 10.1016/j.neuroscience.2006.09.004. Epub 2006 Oct 19. Neuroscience. 2007. PMID: 17055652 Review.
-
News from the protein mutability landscape.J Mol Biol. 2013 Nov 1;425(21):3937-48. doi: 10.1016/j.jmb.2013.07.028. Epub 2013 Jul 26. J Mol Biol. 2013. PMID: 23896297 Review.
Cited by
-
Assessment of computational methods for predicting the effects of missense mutations in human cancers.BMC Genomics. 2013;14 Suppl 3(Suppl 3):S7. doi: 10.1186/1471-2164-14-S3-S7. Epub 2013 May 28. BMC Genomics. 2013. PMID: 23819521 Free PMC article.
-
Human allelic variation: perspective from protein function, structure, and evolution.Curr Opin Struct Biol. 2010 Jun;20(3):342-50. doi: 10.1016/j.sbi.2010.03.006. Curr Opin Struct Biol. 2010. PMID: 20399638 Free PMC article. Review.
-
SIFT web server: predicting effects of amino acid substitutions on proteins.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W452-7. doi: 10.1093/nar/gks539. Epub 2012 Jun 11. Nucleic Acids Res. 2012. PMID: 22689647 Free PMC article.
-
Challenges Related to the Use of Next-Generation Sequencing for the Optimization of Drug Therapy.Handb Exp Pharmacol. 2023;280:237-260. doi: 10.1007/164_2022_596. Handb Exp Pharmacol. 2023. PMID: 35792943
-
Elucidating the genotype-phenotype relationships and network perturbations of human shared and specific disease genes from an evolutionary perspective.Genome Biol Evol. 2014 Oct 5;6(10):2741-53. doi: 10.1093/gbe/evu220. Genome Biol Evol. 2014. PMID: 25287147 Free PMC article.
References
-
- Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. - PubMed
-
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. - PubMed
-
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

